hanno-poster-PCBworkshop_PCB
advertisement
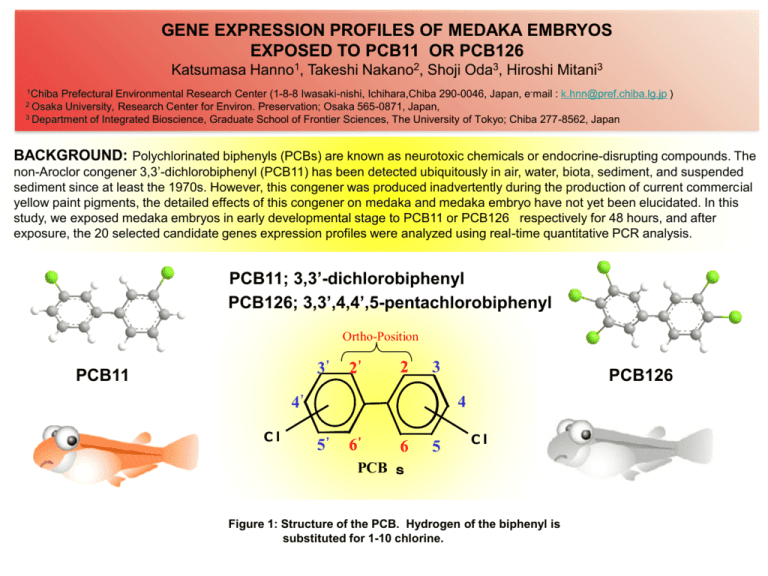
GENE EXPRESSION PROFILES OF MEDAKA EMBRYOS EXPOSED TO PCB11 OR PCB126 Katsumasa Hanno1, Takeshi Nakano2, Shoji Oda3, Hiroshi Mitani3 1Chiba Prefectural Environmental Research Center (1-8-8 Iwasaki-nishi, Ichihara,Chiba 290-0046, Japan, e-mail : k.hnn@pref.chiba.lg.jp ) University, Research Center for Environ. Preservation; Osaka 565-0871, Japan, 3 Department of Integrated Bioscience, Graduate School of Frontier Sciences, The University of Tokyo; Chiba 277-8562, Japan 2 Osaka BACKGROUND: Polychlorinated biphenyls (PCBs) are known as neurotoxic chemicals or endocrine-disrupting compounds. The non-Aroclor congener 3,3’-dichlorobiphenyl (PCB11) has been detected ubiquitously in air, water, biota, sediment, and suspended sediment since at least the 1970s. However, this congener was produced inadvertently during the production of current commercial yellow paint pigments, the detailed effects of this congener on medaka and medaka embryo have not yet been elucidated. In this study, we exposed medaka embryos in early developmental stage to PCB11 or PCB126 respectively for 48 hours, and after exposure, the 20 selected candidate genes expression profiles were analyzed using real-time quantitative PCR analysis. PCB11; 3,3’-dichlorobiphenyl PCB126; 3,3’,4,4’,5-pentachlorobiphenyl Ortho-Position 3' PCB11 2' 2 3 4' Cl PCB126 4 5' 6' 6 5 Cl PCB s Figure 1: Structure of the PCB. Hydrogen of the biphenyl is substituted for 1-10 chlorine. MATERIALS AND METHODS Medaka embryos and exposure to PCB11 or PCB126: Fertilized eggs and embryos were obtained from natural mating of adult orange-red strain medaka fish originally derived from the colony kept in The University of Tokyo and maintained at 26±0.5 oC under a 14 h-light and 10 h-dark cycle. Newly fertilized eggs (200-300 embryos) were placed in a 100 ml beaker with distilled water and incubated for 24 hours at 26±0.5 oC to develop. On the next day, live embryos (stage17) were settled into wells of microtiter plates filled with 200l of waterborne vehicle (DMSO) with 10, 50, 100g/L of PCB11 or 1g/L of PCb126. Total RNA isolation and real-time PCR experiments :. Total RNA was isolated from the exposed embryos by ISOGEN (Nippon Gene Co., Tokyo, Japan) and then purified with RNeasy Mini Kit (QIAGEN). cDNAs were synthesized from 1 g of total RNA using Rever TraAce-TM (Toyobo, Osaka, Japan) and oligo(dT)20 primers. The relative amounts of the transcripts were quantified using real-time PCR which was performed using SYBR® Premix Ex TaqTM in a Smart Cycler®II System (Takara, Shiga, Japan) and specific primers (Hanno et al., in preparation), according to the manufacturer’s instructions. For each sample, gene expression was analyzed in triplicate with the following protocol: 95 oC for 10 s, followed by 45 cycles of 95 oC for 5 s and 60 oC for 20 s. At the completion of each PCR run, the obtained PCR products were subjected to melting curve analysis to ensure that only a single product was amplified. At least three technical replicates of each RNA sample were conducted. Expression data were quantified based on threshold cycle (Ct) values. For each gene, the Ct values for each sample were averaged and normalized to the mean Ct of EF-1which is expressed ubiquitously in tissues and widely used as gene expression control.. Medaka (Oryzias latipes ) Small teleosts such as Medaka or Zebrafish are established as model organisms in risk evaluation of environmental contaminants, however, little is known about molecular machineries of dioxin reception and toxicity expression in nonmammalian species and we do not know whether we can apply the TEQ concept to fish system as well as mammalian system. Recently, a draft genome of Medaka (Oryzias latipes) was released and 4 AhR genes were found in Medaka genome, while 3 AhRs are known for the other nonmammalian genomes and only 1 AhR in mammals. . Method of egg collection and observation ② Medaka pair (♂1-♀1or2) ① Water tanks breeding Medaka ⑥ Observe the embryos under stereo-microscope. ⑦RNA extraction ③ Collect fertilized eggs every morning. ⑤ Settle the eggs into each well of a microtiter plate wit h 200m l of dioxin solution. ⑧Quantitative RT- PCR ④ Incubate the eggs in an incubator for 24 hours at 26±0.5℃. On the next day , remove dead eggs. Table1: The 20 genes for biomarker on the evaluation of environmental water. (bio-marker genes) Group 1 2 No. Biomaker symbol 1 CYP1A1 2 UDPGT 3 AhR1b-1 4 AhR2a 5 AhRR 6 ER-β 7 CACHD1 8 RAR-α 9 ER-α 10 VEGF-R 11 3 12 13 14 Gene ID (Ensemble Gene ID) cp1a1 (ENSORLG00000014421) ENSORLG00000013131 ahr (ENSORLG00000000137) ENSORLG00000000135 AHRR (ENSORLG00000005309) Q8UW75_ORYLA (ENSORLG00000017721) CACHD1 (ENSORLG00000010145) RARA (ENSORLG00000004347) ESR1 (ENSORLG00000014514) ENSORLG00000001940 AGXT(2 of 2) (ENSORLG00000016389) MTF1 MTF1 (ENSORLG00000014411) Tropomyosin ENSORLG00000007803 HSP90AA1(2 of 2) HSP90 (ENSORLG00000017525) AGXT Gene title Intercellular reaction Cytochrome P450 1A detoxication metabolism UDP glucuronosyltransferase xenobiotic metabolism Aryl Hydrocarbon Receptor 1 arylhydrocarbon receptor Aryl Hydrocarbon Receptor 2 arylhydrocarbon receptor AhR Repressor arylhydrocarbon receptor repressor Estrogen receptor beta cell proliferation VEGFR angiogenesis cell proliferation angiogenesis bone differentiation reproduction function cell proliferation vascular endothelial cell function Alanin-glyoxylate aminotransferase hepatocellular proliferation Cache domain-containing protein 1 Retinoic acid receptor alpha Estrogen receptor Tropomyosin metal-response homeostasis muscle-contraction regulation Heat shock protein hsp90 transcriptional regulation transcriptional regulation Metal-response transcription factor1 15 TBP TBP (ENSORLG00000010757) TATA-box binding protein 16 TNF-R ENSORLG00000016219 TNF receptor 17 CDC37 18 HSP70 19 MT 20 Ependymin1 4 CDC37(1 of 2) (ENSORLG00000005543) HSP71_ORYLA (ENSORLG00000000233) MT_ORYLA (ENSORLG00000015580) EPDR1 (ENSORLG00000006266) Hsp90 co-chaperone Cdc37 Heat shock 70 kDa protein 1 (HSP70-1) metallothionein Ependymin related protein 1 inflammatory supression immunological control protein stabilization folding anti-cell death anti-inflammatory homeostasis nervous system heavy metal poisoning neuroplasticity angiogenesis We classified the biomarker genes into four groups, using a medaka customized microarray : Group1 (CYP1A1, UDPGT, AhR1b-1, AhR2a, AhRR, ER) ; detoxication metabolism genes, Group2 (CACHD1, RAR-, ER-, VEGF-R) ; endocrine / reproduction genes, Group3 (AGXT, MTF1, Tropomyosin, HSP90) ; cell proliferation genes, Group4 (TBP, TNF-R, CDC37, HSP70, MT, Ependymin) ; immunity / nervous system genes, considering the physiological functions and induction by chemicals (Hanno et al., in preparation). In particular, Group1 genes were highly inducible by dioxins and dioxin-like compounds exposure. RESULTS AND DISCUSSION Relative Quantity Chart (CYP1A1) 1000 1. Group1 (CYP1A1, AhRR) Cytochrome P450 1A1 (CYP1A1) and aryl hydrocarbon receptor repressor (AhRR) genes are well known to be highly inducible by dioxin and dioxin-like compounds exposure. Both genes are regulated through the aryl hydrocarbon receptor (AhR)-mediated pathway, which is ubiquitously functional in both mammalian tissues and medaka (Oryzia latipes) embryos. CYP1A1 100 Relative Quantity (Crossing Point) Induced expression of biomarker genes mRNA in medaka embryos exposed to PCB11, PCB126 and 2,3,7,8-T4CDD 10 1 0.1 BL (DMSO 0.1%) PCB11_10ug/L (1) CYP1A1 (2) AhRR The relative expression of AhRR was 3.0 fold induction over vehicle-control by PCB126 or 2,3,7,8-T4CDD exposure, but PCB11 induced little. These results suggested that the ecotoxcities of PCB11 might not be strong as those of PCB126 and 2,3,7,8-T4CDD with high TEF (each TEF value was 0.1 and 1), and PCB11 might have another unknown toxic mechanisms. Further assessment was necessary to make clear the potential ecotoxicities of PCB11. PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure 1. CYP1A1 mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of CYP1A1 mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). Relative Quantity Chart (AhRR) 4.0 AHRR 3.0 Relative Quantity (Crossing Point) The relative expression of CYP1A1 in medaka embryos was highly related to the WHO-TEF (the toxic equivalent factor) for mammals (Hanno, et al., 2010). PCB126 (TEF=0.1) and 2,3,7,8-T4CDD (TEF=1) induced CYP1A1 expression 35 or 330 fold over respectively, but PCB11 (10g/L , 50g/L, 100g/L) induced a little . PCB11_50ug/L 2.0 1.0 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure2. AhRR mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of AhRR mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). RESULTS AND DISCUSSION Relative Quantity Chart (CACHD1) 2.5 CACHD1 contains two cache domains and a VWFA (von Willebrand factor A) domain which is known to be involved in hemostasis (Anantharaman, et al., 2000). This gene was induced by the exposure to heavy-metals or PCBs with low TEF, but was suppressed by PCDDs with high TEF (e.g. 2,3,7,8T4CDD). Retinoic acid (RA), the active derivative of vitamin A. The lack or excess of RA result in developmental malformation on vascular, blood vessel and bone formation (Sharon et al., 2000) (1) CACHD1 The relative expression of CACHD1 was induced by PCB11 exposure. The induced expression level of CACHD1 exposed to PCB11 was 1.5 - 2.0 fold induction over vehicle-control, while PCB126 or 2,3,7,8-T4CDD didn’t. (2) RAR- The relative expression of RAR- was induced by PCB11 and PCB126 exposure, while 2,3,7,8-T4CDD didn’t. Relative Quantity (Crossing Point) 2. Group2 (CACHD1, RAR-) CACHD1 2.0 1.5 1.0 0.5 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure3. CACHD1 mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of CACHD1 mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). Relative Quantity Chart (RAR- ) 2.0 RAR- 1.5 Relative Quantity (Crossing Point) Induced expression of biomarker genes mRNA in medaka embryos exposed to PCB11, PCB126 and 2,3,7,8-T4CDD 1.0 0.5 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure4. RAR- mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of RAR- mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). RESULTS AND DISCUSSION Relative Quantity Chart (HSP90) 1.5 Induced expression of biomarker genes mRNA in medaka embryos exposed to PCB11, PCB126 and 2,3,7,8-T4CDD Relative Quantity (Crossing Point) HSP90 1.0 HSPs (heat shock proteins) are stress-defenssive proteins and are highly induced in response to stresses caused by changes in environmental factors such as high temperature, heavy metal administrations, reactive oxygen production, salinity, etc (Beckmann, et al.,1990) and protect the structure and function of proteins and cells from damages, maintaining the cellular homoeostasis. Tropomyosin is an actin-binding protein that regulates actin mechanics. It is important, among other things, for muscle contraction. (1) HSP90 The relative expression of HSP90 was suppressed by PCB126 or 2,3,7,8-T4CDD, and PCB11 didn’t induce. (2) Tropomyosin The relative expression of Tropomyosin was induced by PCB11 at fifty g/L concentration, and was not induced by PCB126 or 2,3,7,8-T4CDD. 0.5 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure5. HSP90 mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of HSP90 mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). Relative Quantity Chart (Tropomyosin) 3.0 2.5 Tropomyosin Relative Quantity (Crossing Point) 3. Group3 (HSP90, Tropomyosin) 2.0 1.5 1.0 0.5 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure6. Tropomyosin mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of Tropomyosin mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). RESULTS AND DISCUSSION MT Relative Quantity (Crossing Point) Induced expression of biomarker genes mRNA in medaka embryos exposed to PCB11, PCB126 and 2,3,7,8-T4CDD Relative Quantity Chart (Metallothionein, MT) 1.5 1.0 Metallothionein (MT) is a protein that has a high cysteine content, a low molecular mass about 7 kDa, and a high affinity for metals. It binds metals and regulates the homeostasis of essential trace metals such as copper and zinc, also taking a part in counteracting the toxic effects of heavy metals such as cadmium, mercury, and silver (Choi, et al., 2007). Ependymin is a glycoprotein associated with the consolidation of long-term memory, claims involving protection from strokes, and neuronal regeneration. (1) Metallothionein (MT) The relative expression of Metallothionein was suppressed by PCB126, 2,3,7,8-T4CDD and PCB11(100ug/L). PCB11 (50ug/L) induced a little. (2) Ependymin The relative expression of Ependymin was induced by PCB11 exposure. The induced expression level of Ependymin exposed to PCB11 was about 2.0 fold induction over vehicle-control, while PCB126 or 2,3,7,8-T4CDD induced a little. 0.5 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure7. MT mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of MT mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). Relative Quantity Chart (Ependymin) 2.5 Ependymin 2.0 Relative Quantity (Crossing Point) 4. Group4 (Metallothionein, Ependymin) 1.5 1.0 0.5 0.0 BL (DMSO 0.1%) PCB11_10ug/L PCB11_50ug/L PCB11_100ug/L PCB126_1ug/L 2378T4CDD_1ug/L Target-Sample Figure8. Ependymin mRNA expression in medaka embryos exposed to PCB11 and PCB126. The amount of Ependymin mRNA was measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure, the error bars represents SD (n=3). RESULTS AND DISCUSSION PCB126 (1ug/L) or 2,3,7,8-T4CDD (1ug/L) induced CYP1A1 in proportion to its TEF values, AhR2a and AhRR (Group1), and suppressed the genes of Group3 and Group4. On the other hand, PCB11 (10, 50, 100 ug/L) induced CYP1A1 (Group1), CACHD1, RAR- (Group2), and Tropomyosin (Group3). The relative mRNA expressions of these genes were presented in radar figure of 20 square. (Figure9) CYP1A1 /5 Ependymin1 5 UDPGT MT AhR1b-1 4 HSP70 CDC37 TNF-R 3 2 1 0 AhR2a AhRR ER-β TBP CACHD1 HSP90a RAR-α Tropomyosin MTF1 ER-α VEGF-R CYP1A1 Ependymin1 5 MT 4 HSP70 CDC37 TNF-R 3 2 1 0 AhR2a AhRR ER-β TBP CACHD1 HSP90a RAR-α Tropomyosin MTF1 ER-α VEGF-R AGXT AGXT PCB126_1ug/L UDPGT AhR1b-1 2378T4CDD_1ug/L PCB11_10ug/L PCB11_100ug/L PCB11_50ug/L Figure 9. The biomarker genes mRNA expressions in medaka embryos exposed to PCB11, PCB126 and 2,3,7,8-T4CDD, presented in radar figure of 20 square. The amount of those mRNA were measured by quantitative real-time PCR and normalized to the solvent control (0.1% DMSO) for 48 h exposure. (n=3) CONCLUSIONS We have evaluated the ecotoxicities of wastewaters by investigating the biomarker genes expression profiles in the early developmental stage of medaka embryos exposed to wastewaters containing dioxins and dioxin-like compounds. In this study, we evaluated the toxicities of PCB11, obtaining the following conclusions : -1. PCB11 (10, 50, 100 ug/L) induced CYP1A1 (Group1), CACHD1, RAR- (Group2), and Tropomyosin (Group3). -2. PCB126 (1ug/L) and 2,3,7,8-T4CDD (1ug/L) induced CYP1A1 in proportion to its TEF values, AhR2a, AhRR (Group1), and suppressed the genes of Group3 and Group4. -3. The ecotoxcities of PCB11 might not be strong as those of PCB126 and 2,3,7,8-T4CDD with high TEF (each TEF value was 0.1 and 1). -4. PCB11 might have another unknown toxic mechanisms. -5. Further assessment was necessary to make clear the potential ecotoxicities of PCB11. REFERENCES Besselin Anantharaman V, Aravind L. (2000) Trends Biochem Sci. 25(11), 535-537 Beckmann R.P., Mizzen L.E., Welch W.J. (1990) Science. 248, 850-854. Choi C.Y., An K.W., Nelson E.R., Habibi H.R. (2007) Comp. Biochem. Physiol. C145, 595-600. Hahn ME., Allan LL., Sherr DH. (2009) Biochem, Pharmacol. 77, 485-497. Hanno K, Oda S, and Mitani H. (2010) Chemosphere 78, 830-839. Sharon A., Peter J., Luigi M.D.L. (2000) Physiol. Rev. 80, 1021-1054. Weber R, Hagenmaier H (1999); Chemosphere 38 (3):529-549. E-mail is as follows; k.hnn@pref.chiba.lg.jp ( Katsumasa HANNO )