SAS® Global Forum 2014
advertisement

SAS® Global Forum 2014
March 23-26
Washington, DC
Got Randomness?
SASTM for Mixed
and Generalized
Linear Mixed Models
David A. Dickey
NC State University
TM SAS
and its products are the registered
trademarks of SAS Institute, Cary, NC
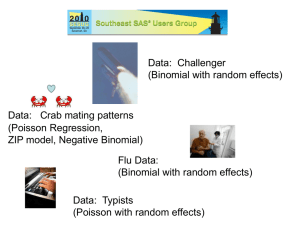
Data: Challenger
(Binomial with
random effects)
Data: Ships
(Poisson with
offset)
Data: Crab mating patterns (X rated)
(Poisson Regression, ZIP model,
Negative Binomial)
Data: Typists
(Poisson with
random effects)
Data: Flu samples
(Binomial with
random effects)
2
“Generalized” non normal distribution
1/3
Like Dislike
Pr{ Like }=2/3
0
.27
1
2
.001
l=0.4055
2/3
.007
Poisson for counts: Y in {0,1,2,3,4, ….}
Mean count l Variance l
Pr{Y=j} = exp(- l )(lj)/(j!)
Link: L = log(l)
Range (over all L): l >0
2/3
.055
Binary for probabilities: Y=0 or 1
Mean E{Y}=p
Variance p(1-p)
Pr{Y=j}= pj(1-p)(1-j)
Link: L=ln(p/(1-p)) = “Logit”
Range (over all L): 0<p<1
3
4
5
3
Mixed (not generalized) Models:
Fixed Effects
and
Random Effects
4
Fixed?
Replication :
Same levels
or
Random?
Different levels
Inference for: Only Observed
Levels
Population of
Levels
Levels :
Picked on
Purpose
Picked at
Random
Inference on:
Means
Variances
Example:
Only These
Drugs
Only These
Fertilizers
All Doctors
All Clinics
All Farms
All Fields
Example:
5
Generalized (not mixed) linear models.
Use link L = g(E{Y}), e.g. ln(p/(1-p)) = ln(E{Y}/(1-E{Y})
Assume L is linear model in the inputs with fixed effects.
Estimate model for L, e.g. L=g(E{Y})=bo + b1 X
Use maximum likelihood
Example:
L = -1 + .18*dose
Dose = 10, L=0.8,
p=exp(0.8)/(1+exp(0.8))=
“inverse link” = 0.86
6
Challenger was mission 24
From 23 previous launches we have:
6 O-rings per mission
Y=0 no damage, Y=1 erosion or blowby
p = Pr {Y=1} = f{mission, launch temperature)
Features: Random mission effects
Logistic link for p
proc glimmix data=O_ring;
class mission;
model fail = temp/dist=binomial s;
random mission;
run;
Generalized
Mixed
8
Estimated G matrix is not positive definite.
Covariance Parameter Estimates
Cov
Parm
mission
Estimate
2.25E-18
Standard
Error
.
We “hit the boundary”
Solutions for Fixed Effects
Effect
Intercept
temp
Estimate
Error
DF
t Value
Pr > |t|
5.0850
-0.1156
3.0525
0.04702
21
115
1.67
-2.46
0.1106
0.0154
9
Likelihood
2
M
0
E2 0
E2 0
2
M
0
10
Just logistic regression – no mission variance component
11
Empirical Logit
Flu Data CDC
Active Flu Virus
Weekly Data % positive
% positive
data FLU;
input fluseasn year t week pos specimens;
pct_pos=100*pos/specimens;
logit=log(pct_pos/100/(1+(pct_pos/100)));
label pos = "# positive specimens";
label pct_pos="% positive specimens";
label t = "Week into flu season (first = week 40)";
label week = "Actual week of year";
label fluseasn = "Year flu season started";
12
(1) GLM all effects fixed
(harmonic main effects
insignificant)
“Sinusoids”
S(j) = sin(2pjt/52)
C(j)=cos(2pjt/52)
PROC GLM DATA=FLU;
class fluseasn;
model logit = s1 c1
fluseasn*s1 fluseasn*c1 fluseasn*s2 fluseasn*c2
fluseasn*s3 fluseasn*c3 fluseasn*s4 fluseasn*c4;
output out=out1 p=p;
data out1; set out1; P_hat = exp(p)/(1+exp(p));
label P_hat = "Pr{pos. sample} (est.)"; run;
13
(2) MIXED analysis
on logits
Random harmonics.
Normality assumed
Toeplitz(1)
2j 0
0 2
j
PROC MIXED DATA=FLU method=ml;
** reduced model;
class fluseasn;
model logit = s1 c1 /outp=outp outpm=outpm ddfm=kr;
random intercept/subject=fluseasn;
random s1 c1/subject=fluseasn type=toep(1);
random s2 c2/subject=fluseasn type=toep(1);
random s3 c3/subject=fluseasn type=toep(1);
random s4 c4/subject=fluseasn type=toep(1); run;
14
(3) GLIMMIX
analysis
Random harmonics.
Binomial assumed
(overdispersed –
lab effects?)
PROC GLIMMIX DATA=FLU; title2 "GLIMMIX Analysis";
class fluseasn;
model pos/specimens = s1 c1 ; * s2 c2 s3 c3 s4 c4;
random intercept/subject=fluseasn;
random s1 c1/subject=fluseasn type=toep(1);
random s2 c2/subject=fluseasn; ** Toep(1) - no converge;
random s3 c3/subject=fluseasn type=toep(1);
random s4 c4/subject=fluseasn type=toep(1);
random _residual_;
covtest glm;
output out=out2 pred(ilink blup)=pblup
pred(ilink noblup)=overall pearson = p_resid; run;
15
output out=out2
pred(ilink blup)=pblup
pred(ilink noblup)= overall
pearson = p_resid;
run;
Pearson Residuals:
Used to check fit when using
default (pseudo-likelihood)
Variance should be near 1
proc means mean var;
var p_resid; run;
Without random _residual_; variance 3.63.
With random _residual_; variance 0.83.
--------------------------------------------------------------Fit Statistics
-2 Res Log Pseudo-Likelihood
341.46
Generalized Chi-Square
1707.29
Gener. Chi-Square / DF
4.59
-------------------------------------------------------------------------
Or… use method=quad
16
Type III Tests of Fixed Effects
Effect
S1
c1
Output due to
covtest glm;
Label
Independence
Num
DF
1
1
Den
DF
8
8
F Value
34.93
25.49
Pr > F
0.0004
0.0010
Tests of Covariance Parameters
Based on the Residual Pseudo-Likelihood
DF
6
-2 Res Log P-Like
1312.34
ChiSq
970.88
Pr > ChiSq
<.0001
Note
MI
MI: P-value based on a mixture of chi-squares
random _residual_ does not affect
the fit (just standard errors)
17
Could try 2 parameter Beta distribution instead:
PROC GLIMMIX DATA=FLU; title2 "GLIMMIX Analysis";
class fluseasn;
model f = s1 c1 /dist=beta link=logit s;
random intercept/subject=fluseasn;
random s1 c1/subject=fluseasn type=toep(1);
random s2 c2/subject=fluseasn type=toep(1);
random s3 c3/subject=fluseasn type=toep(1);
random s4 c4/subject=fluseasn type=toep(1);
output out=out3 pred(ilink blup)=pblup
pred(ilink noblup)=overall
pearson=p_residbeta; run;
18
Without BLUPS
Binomial Assumption
and
with BLUPS
19
Without BLUPS
Beta Assumption
and
with BLUPS
20
Poisson Example: Wave induced damage
incidents in 40 ships (ship groups)
Variables:
Factorial Effects (fixed, classificatory):
Ship Type
5 levels A,B,C,D,E
Year Constructed
4 levels
Years of Operation 2 levels
Constructed
Operated
60-64
65-69
70-74
60-74
ABCDE
ABCDE
ABCDE
75-79
ABCDE
ABCDE
ABCDE
75-79
-XABCDE
Covariate (“offset” - continuous) =
Time in service (“Aggregate months”)
Incidents (dependent, counts)
Source” McCullough & Nelder
(but I ignore cases where year
constructed > period of operation)
21
proc glimmix data=ships;
Title "Ignoring Ship Variance";
class operation construct shiptype;
model incidents = operation construct shiptype/
dist=poisson s offset=log_service; run;
ln(service)
Poisson: ln(l) – ln(service) = b0 + b1(operation) + b2(construct) + b3(ship_type)
ln(l/service)
-2 Log Likelihood
(more fit statistics)
Pearson Chi-Square
Pearson Chi-Square / DF
136.56
>1
Ship variance?
42.28
1.69
22
Without ship variance component:
Type III Tests of Fixed Effects
Effect
Operation
Construct
Shiptype
Num
DF
Den
DF
F Value
Pr > F
1
3
4
25
25
25
10.57
9.72
6.50
0.0033
0.0002
0.0010
23
PROC GLIMMIX data=ships method=quad;
class operation construct shiptype ship;
model incidents = operation construct shiptype/
dist=poisson s offset=log_service;
covtest "no ship effect" glm;
* random ship;
random intercept / subject = operation*construct*shiptype;
run;
Covariance Parameter Estimates
Cov Parm
Intercept
Subject
Operat*Constr*Shipty
Estimate
0
Standard
Error
.
24
Fit Statistics
-2 Log Likelihood
136.56
Fit Statistics for Conditional Distribution
-2 log L(incidents | r. effects)
Pearson Chi-Square
Pearson Chi-Square / DF
136.56
42.27
1.24
Type III Tests of Fixed Effects
Effect
Operation
Construct
shiptype
Num
DF
1
3
4
Den
DF
25
25
25
F Value
10.57
9.72
6.50
Tests of Covariance Parameters
Based on the Likelihood
Label
DF
-2 Log Like
no ship effect
1
136.56
MI: P-value based on a mixture of chi-squares.
ChiSq
.
Pr > F
0.0033
0.0002
0.0010
no
changes
covtest "no ship effect" glm;
Pr > ChiSq
1.0000
Note
MI
25
Horseshoe Crab study
(reference: SAS GLIMMIX course notes):
Female nests have “satellite” males
Count data – Poisson?
Generalized Linear
Features (predictors):
Carapace Width, Weight, Color, Spine condition
Random Effect: Site
Mixed Model
To nest
Go
State
26
proc glimmix data=crab;
class site;
model satellites = weight width /
dist=poi solution ddfm=kr;
random int / subject=site;
output out=overdisp pearson=pearson; run;
proc means data=overdisp n mean var;
var pearson; run;
N
Mean
Variance
--------------------------173 -0.0258264
2.6737114
---------------------------
proc univariate data=crab normal plot;
var satellites; run;
Fit Statistics
Gener. Chi-Square / DF
2.77
Cov Parm
Intercept
Subject Estimate
site
0.1625
Effect
Intercept
weight
width
Estimate
-1.1019
0.5042
0.0318
Pr > |t|
0.2527
0.0035
0.5229
Histogram
15.5+*
.*
.
12.5+*
.*
.**
9.5+**
.***
.**
6.5+*******
.********
.**********
3.5+**********
.*****
Zero Inflated ?
.********
0.5+*******************************
----+----+----+----+----+----+* may represent up to 2 counts
# Boxplot
1
0
1
0
1
1
3
3
6
4
13
15
19
19
9
16
62
|
|
|
|
|
|
|
+-----+
|
|
|
|
*--+--*
|
|
+-----+
27
Zero Inflated Poisson (ZIP)
p0 + (1 p0 )e l l0 / 0! p0 + (1 p0 )e l for j 0
Pr{Y j}
l j
(
1
p
)
e
l / j!
for j 0
0
E{Y } (1 p0 )l
Q: Can zero inflation cause overdispersion (2>)?
Recall: in Poisson, 2=l
28
E{Y } (1 p0 )l
E{Y (Y 1)} (1 p0 ) j ( j 1)e l l j / j!
j 1
(1 p0 )l2 e l l j 2 /( j 2)! (1 p0 )l2 E{Y 2 }
j 2
E{Y 2 } (1 p0 )l2 + (1 p0 )l
2 E{Y 2 } ( EY ) 2 (1 p0 )(l2 + l ) l2 (1 p0 ) 2
(1 p0 )l + l2 p0 (1 p0 ) + l2 p0 (1 p0 )
2
A: yes!
29
Nice job Grandpa.
That proof just about
put everyone to sleep
30
Zero Inflated Poisson - (ZIP code )
Dickey
ncsu
SAS Global Forum
Washington DC
20745
proc nlmixed data=crab;
parms b0=0 bwidth=0 bweight=0 c0=-2 c1=0 s2u1=1 s2u2=1;
x=c0+c1*width+u1; p0 = exp(x)/(1+exp(x)); * width affects p0;
eta= b0+bwidth*width +bweight*weight +u2;
lambda=exp(eta);
if satellites=0 then
loglike = log(p0 +(1-p0)*exp(-lambda));
else loglike =
log(1-p0)+satellites*log(lambda)-lambda-lgamma(satellites+1);
expected=(1-p0)*lambda; id p0 expected lambda;
model satellites~general(loglike);
Random U1 U2~N([0,0],[s2u1,0,s2u2]) subject=site;
predict p0+(1-p0)*exp(-lambda) out=out1; run;
31
Parameter Estimates
Parameter
Variance
for
p0
l
b0
bwidth
bweight
c0
c1
s2u1
s2u2
Estimate
2.7897
-0.0944
0.4649
13.3739
-0.5447
0.5114
0.1054
t
2.55
-1.65
2.38
4.42
-4.61
1.12
1.67
Pr>|t|
0.0268
0.1267
0.0366
0.0010
0.0008
0.2852
0.1239
Lower
Upper
0.3853 5.1942
-0.2202 0.0314
0.0347 0.8952
6.7078 20.0401
-0.8049 -0.2844
-0.4905 1.5133
-0.0339 0.2447
weight
affects l
width
affects p0
32
From fixed part of model, compute
Pr{count=j} and plot (3D) versus
Weight, Carapace width
33
34
35
Your talk
seems better
now Grandpa!
Another possibility: Negative binomial
Number of failures until kth success ( p=Prob{success} )
k + j 1 k 1
k + j 1 k
p (1 p) j p
p (1 p) j
Pr{Y j}
k 1
k 1
k (1 p)
k
, p
p
k+
2
l 2 l + 1 l2 l
k
p
(k + )
k
as 1 / k 0
44
Negative binomial:
In SAS, k (scale) is our 1/k
proc glimmix data=crab; class site;
model satellites = weight width / dist=nb solution ddfm=kr;
random int / subject=site; run;
Fit Statistics
-2 Res Log Pseudo-Likelihood
Generalized Chi-Square
Gener. Chi-Square / DF
539.06
174.83
1.03
Covariance Parameter Estimates
Cov Parm
Intercept
Scale
Effect
Intercept
weight
width
Estimate
-1.2022
0.6759
0.01905
Subject
site
Standard
Error
1.6883
0.3239
0.08943
Estimate
0.09527
0.7659
DF
168.5
156.6
166.2
Std. Error
0.07979
0.1349
t Value
-0.71
2.09
0.21
Pr > |t|
0.4774
0.0386
0.8316
45
Population average model vs. Individual Specific Model
8 typists
Y=Error counts (Poisson distributed)
ln(li)= ln(mean of Poisson) = +Ui for typist i so li=e+Ui
conditionally (individual specific)
Distributions for Y, U~N(0,1) and =1
l=e=e1=2.7183
= mean for
“typical” typist
(typist with U=0)
Population average model
Expectation ||||| | | of individual distributions averaged
across population of all typists.
Run same simulation for 8000 typists, compute mean
of conditional population means, exp(+U).
The MEANS Procedure
Variable
N
Mean
Std Dev Std Error
ƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒ
lambda
8000 4.4280478 6.0083831 0.067175
ƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒƒ
Z=(4.428-2.7183)/0.06718 = 25.46 !!
Population mean is not e
Conditional means, +U, are lognormal.
Log(Y)~N(1,1)
E{Y}=exp(+0.52) = e1.5 = 4.4817
47
Main points:
1. Generalized linear models with random effects are
subject specific models.
2. Subject specific models have fixed effects that
represent an individual with random effects 0
(individual at the random effect distributional means).
3. Subject specific models when averaged over the
subjects do not give the model fixed effects.
4. Models with only fixed effects do give the fixed effect
part of the model when averaged over subjects and
are thus called population average models.
48