14. Cellular Reproduction

Gabriel Krotkov
XV. Cellular Reproduction
Biology Notes
3/18/14
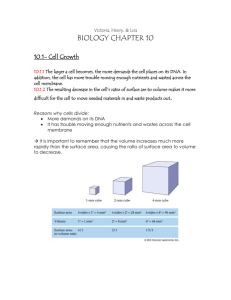
A. Cell size limit/Surface : Volume
1. Cells reproduce when cell size is too large.
2. Diffusion as a method for cells a. Slow + Inefficient over the long distances that a large cell creates
3. DNA a. A nucleus cannot make proteins fast enough to keep up
4. Surface Area : Volume a. SA increases less than volume b. SA gets outpaced by volume growth c. SA supplies volume w/o surface area, volume cannot be.
5. Example a. Cube like so b. SA:6, V:1 c. At 2, instead of 1 3:1 d. As you see, quickly outgrows SA
B. Purposes for cell division
1. Growth
2. Replace old cells
3. Reproduce
C. Cell Cycle (see diagram 1)
1. Period of time from cell creation to when it divides
2. Eukaryotic Phases a. Interphase = prep b. Mitosis – nuclear division c. Cytokinesis – Division of Cytoplasm
3. Restriction point a. A point of no return b. After G1, before S.
4. Prokaryotic Phases a. Growth b. Chromosome split to either side b. Septum grows to split the two c. Cell splits along Septum
D. Interphase
1. Three sections a. G1 b. S c. G2
2. G1 a. Cell growth b. Normal acticity
3. G0 a. Part of G1, but for cells that don’t divide b. G1 holding pattern
4. Synthesis a. DNA Synthesis b. Makes a new copy of the chromosones c. S is to Mitosis as G2 is to Cytokinesis d. Genetic Material for daughter cells are created e. Each strand serves as a model to create a new DNA molecule. f. Semi-conservative g. Steps
i. Helicase unzips the DNA strand, beginning at the origins of replication, short stretches of DNA that have a specific sequence of nucleotides. Note that rather than a single helicase unzipping an entire DNA strand, multiple helicases open
“replication bubbles”, which are worked on simultaneously to speed up replication. strand.
ii. SSBP’s (single strand binding proteins) stabilize and hold open the DNA
iii. RNA Primase creates a leading group of RNA about 10 RNA bases long
(complementary to the beginning of the sequence). Leading strand only needs one primer, but the lagging strand needs 1 primer per okazaki fragment.
iv. RNA Polymerase III creates DNA nitrogen bases from the primase bases to continue on
v. On the strand going 5’ to 3’ (lagging strand), the parts are built in sections, called Okazaki fragments. An RNA primer is set up and the polymerase III moves in as in the leading strand in order to fill the section after the primer with
DNA bases.
vi. Polymerase I treats the primer RNA from primase, turning it into DNA by replacing U with T and removing an oxygen.
vii. Ligase seals the Okazaki fragments
viii. SSBP’s leave, and the DNA reseals. h. Interesting point: each DNA strand does not have to be copied base by base; often multiple portions of the strand will be “opened” and worked on by proteins system at one time. i. Antiparallel elongation
i. The two strands of DNA in a double helix are antiparallel, meaning that they are oriented in opposite directions to each other (relative to 3’ or 5’ ends)
ii. Because DNA polymerases can only add nucleotides to the 3’ end of a strand rather than the 5’ end, this means that one polymerase in each bubble will continually work 5’ 3’, while the other polymerase will work away from the replication fork. The strand going from 5’ 3’ is called the leading strand, while the strand going from 3’ 5’ is called the lagging strand. See okazaki fragments above.
iii. j. Important distinctions:
i. All proteins that act on DNA in this process are all part of one complex
ii. Proteins don’t move along DNA, DNA moves through proteins. It’s just simpler to understand it the other way.
k. Proofreading/Repairing
i. Polymerases follow each replication bubble and proofread after the 1 st polymerase attaches nucleotides.
ii. Nucleotide excision repair: a DNA repair system in which a nuclease (a
DNA cutting enzyme) removes an error and then polymerase/ligase replaces it. l. Replicating the ends of DNA Molecules
i. In the case of linear DNA molecules, there is a portion of DNA that polymerases cannot replicate or repair; the 5’ end of a daughter DNA strand.
ii. This causes an issue: one strand will get perpetually shorter as cells divide. This is a proposed cause of aging.
iii. To hold off, but not prevent, this eventuality, cells have special nucleotide sequences at the end of each strand: telomeres.
iv. Telomeres have no purpose, but rather are repetitions of a single short nucleotide sequence, allowing the strand to have nucleotides to spare and thus survive many replications intact.
v. Telomerase catalyzes the lengthening of telomeres jn eukaryotic germ cells, and compensates for the shortening that occurs in cell division. This is only applied to cells that must persist virtually unchanged from an organism to its offspring. In humans, this is germ cells that produce gametes.
5. G2 a. Copies organelles b. Cytokinesis to S’s Mitosis
6. Timeline a. In human cells, this is not an unreasonable timeframe:
i. G1 = 5-6 hrs
ii. S = 10-12 hrs
iii. G2 = 4-5 hrs
iv. Mitosis = <1 hr
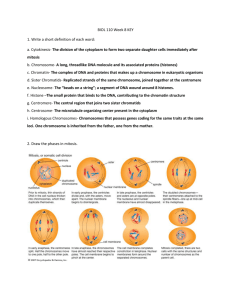
E. Mitosis
1. Division of the nucleus in somatic cells
2. Results in two genetically equivalent daughter cells
3. Steps a. Prophase
i. Prometaphase b. Metaphase c. Anaphase d. Telophase
4. Prophase a. Centrioles move to the sides (only in animal cells) b. Nucleus envelope dissolves c. Spindle Fibers form d. Chromatin condenses into Chromosomes
5. Metaphase a. The spindle fibers move sister chromatids to the center
i. Microtubules connect to chromosomes on opposite sister chromatids, via the kinetochores (called “capturing the kinetochores”)
ii. These microtubules then adjust the chromosomes into the center of the nucleus by opposite pulls: microtubules from both side pull equally, brining the chromosomes into roughly the center.
iii. Asters attach to the envelope and the microtubules that are not connected to kinetochores attach to their coordinating pair on the other side. b. Kinetochore
i. Motor protein structure on centromere
ii. Spindle fibers grab the centromeres via these
6. Anaphase a. Sister chromatids split via spindle fibers
i. Separase first removes the cohesins on each of the sister chromatids
ii. Once the cohesins are removed, the kinetochores, being motor proteins
(dynein arms) walk the chromosomes along the microtubules. Motor proteins at the spindle poles also “reel in” the microtubules, and thus, the chromatids.
iii. The microtubules depolymerize at their kinetochore ends after the motor proteins pass.
iv. As the chromatids get closer to the poles, the nucleus enlongates.
7. Telophase a. Nuclear envelope reforms b. Spindles break down c. Chromosomes uncoil to form chromatin
F. Meiosis
1. Creates gametes a. Haploid cells, meaning 23 chromosomes b. Two gametes = one zygote, total 46 chromosomes
2. Double parent production
3. Germ cells begin Meiosis a. Somatic cell b. They undergo synthesis, 92 chromosomes c. Splits into two of 46 chromosomes d. Splits into 4 of 23 chromosomes
4. Alleles a. A given trait of a chromosome b. Carried on the same chromosome, just a different version
5. Diploid/Haploid cells a. Somatic cells and Germ cells are diploid (2n) b. Gametes are haploid (n) c. Meiosis goes from Diploid to Haploid
6. Process Stages a. Meiosis I
i. Prophase, Metaphase, Anaphase, Telophase I b. Meiosis II
i. Prophase Metaphase Anaphase Telophase II c. Chromosome progression
i. 46 – diploid
ii. 92 – Synthesis
iii. (2) 46 – Meiosis I
iv. (4) 23 – Meiosis II
7. Meiosis I a. Prophase I
i. Chromosomes condense
ii. Chromosomes synapse (connected physically)
iii. Homologs cross over (exchange genetic material)
iv. Nuclear membrane breaks down
v. Spindle forms, and then microtubules from each pole attack to kinetochores of each chromosome.
vi. Crossing over results in chiasmata, X-shaped regions that are created when the two original sister chromatids are still held together by cohesion. b. Metaphase I
i. Tetrads line up randomly along the metaphase plate
ii. Tetrad – 4 chromosomes
iii. Tetrads are brought to the plate by the nearly equal pulling of each fiber, which equalizes at the plate (center of the cell)
iv. Homologs attach to their respective fibers by the kinetochore. c. Anaphase I
i. Homologs separate – Disjunction: caused by breakdown of proteins responsible for chromosomal cohesion
ii. Homologs move towards opposite poles
iii. Sister chromatid cohesion persists at the centromere, causing chromatids to move as a unit toward the same pole d. Telophase I & Cytokinesis
i. Each half of the cell has a complete haploid set of duplicated chromosomes; each composed of two sister chromatids.
ii. Cytokinesis occurs, forming 2 haploid daughter cells
iii. Cleavage furrow for next division is created
iv. In some instances, chromosomes decondense and nuclear envelopes form; occurs if cell will be in form long enough to have use for it.
v. No chromosomal duplication.
8. Meiosis II a. Prophase II
i. Spindle forms
ii. Chromosomes move towards metaphase II plate b. Metaphase II
i. Spindle moves sister chromatids (not genetically identical) towards metaphase II plate
ii. Kinetochores of sister chromatids attach to microtubules from the poles c. Anaphase II
i. Proteins holding together sister chromatids break down
ii. Chromatids move towards respective centrosomes d. Telophase II & Cytokinesis
i. Nuclei form, chromosomes decondense
ii. Cytokinesis occurs as in mitosis
iii. Results in 4 genetically distinct haploid cells
G. Cytokinesis
1. Overview a. Begins half-into mitosis
b. Separates cytoplasm equally between daughter cells
2. Process a. Cleavage
i. A cleavage furrow appears in the cell surface near the metaphase plate
(remember, an imaginary construct)
ii. Actin filaments with myosin associates form a contractile ring that deepens the cleavage furrow until the parent cell is pinched into two identical cells.
H. Organelles and structures associated with cellular reproduction
1. Chromosomes
a. One very long linear DNA molecule associated with proteins.
i. An uncondensed, unduplicated chromosome has a single centromere and two arms.
b. Human somatic cells have 46 chromosomes
c. Chromosomes come in pairs
d. Do not interact during mitosis
e. Each chromosome and its copy stay together
i. Sister chromatids – each ½ of the pair.
ii. Totally identical
iii. ½ of the chromatid is called a “chromatid arm”; these are demarcated by the centromere.
f. Centromere
i. Holds sister chromatids together
ii. Protein complexes called cohesions aid in doing so, creating sister chromatid cohesion along the entire chromatid pair.
g. Made of DNA + Histone proteins h. Function
i. Carries thousands of genes, which specify traits. i. Kinetochore
i. On each sister chromatid, a structure of proteins associated with specific sections of chromosomal DNA at each centromere.
ii. Face in opposite directions
2. Chromatin
a. Unraveled version of chromosones
b. 10,000x longer
c. Cannot be seen under microscope
d. Takes energy to uncoil e. Chromatin is the form DNA takes while the cell is in interphase; because it is more easy to access for use in protein synthesis.
3. Nucleosome
a. DNA wrapped around Histones
b. Stays together via polarity
4. Mitotic Spindle a. Structure
i. Fibers made of microtubules w/proteins
ii. Has a radial array of short microtubules extending from the centrosomes, called Asters. These connect the centrosomes to the nuclear envelope.
iii. Contains chromosomes, which organize microtubules and act as anchors at the end of the nucleus
iv. Contains spindle microtubules that connect centrosomes to chromosomes. They also connect to microtubules from the other centrosome b. Function
i. Take apart duplicated chromosomes by the kinetochore during anaphase c. Mechanism
i. Microtubules connect to chromosomes on opposite sister chromatids, via the kinetochores (called “capturing the kinetochores”)
ii. These microtubules then adjust the chromosomes into the center of the nucleus by opposite pulls: microtubules from both side pull equally, brining the chromosomes into roughly the center.
iii. During metaphase, the asters attach to the envelope and the microtubules that are not connected to kinetochores attach to their coordinating pair on the other side. d. Creation
i. Begins in centrosome, where the microtubules are organized
ii. The centrosome has a pair of centrioles that are located at the center; but these are not necessary for mitosis: in cells w/o them, mitosis proceeds apace.
iii. The microtubules used for the creation of the spindle are disassembled from elsewhere in the cell.
iv. During interphase, centrosome duplicates, and during prophase/metaphase they take part in chromosome organization in the nucleus.
I. Cancer
1. Incorrect genetic information = uncontrolled cell division
2. Cell Cycle has built-in checkpoints in cycle
3. Checkpoint genes a. Code for proteins that monitor condition of DNA, chromosomes, and spindles b. eg. Growth factors c. Can arrest cell cycle
4. Proto-oncogenes a. Regulate cell division and cell cycle b. Determine division speed c. Can become oncogenes
5. Tumor suppressors a. Inhibit cell division
6. Benign Tumor a. Harmless b. Do not spread
7. Malignant Tumors
a. Spreads b. Deadly
8. Metastasis a. Spreads through lymph system of bloodstream
9. 4 characteristics of cancerous tumors a. Grow/Divide abnormally b. Cytoplasm and cell membrane is altered c. Weakened adhesion d. Lethal if untreated
J. Meiosis in Humans
1. Males a. Primary germ cell is spermatocyte b. Makes 4 sperm cells
2. Females a. Oocyte is primary germ cell b. To feed the baby, cytoplasm is divided unevenly c. 1 cell gets all the cytoplasm the other three die d. The 3 that die are called polar bodies e. Polar bodies dispose of genetic material
3. Mating a. Gametes unite, nucleus’ fuse b. 2 haploid nuclei = 1 diploid nuclei c. Unknown why 2 given gametes work
K. Mutation & Chromosomal Mistakes
1. Non-disjunction a. Failure of chromosomes to separate during anaphase (I or II) b. Caused by random mistakes c. Spindle Fibers don’t attach at one side d. Chromosome cannot find homolog
2. Aneuploidy a. Cells with one extra or one less chromosome b. Autosomal Aneuploidy is usually fatal for humans
3. Monosomy a. One missing chromosome b. 45 chromosomes
4. Trisomy a. One extra chromosome c. 47 Chromosomes c. eg. Down Syndrome is trisomy 21
5. Polyploidy a. Three or more of each type of chromosome b. 69 chromosomes – dead human
6. Types of chromosomal mutation a. Deletion – A part of a chromosome is missing b. Insertion – A part of a chromatid breaks off and attaches to its sister chromatid
c. Inversion – piece of a chromatid reinserted backwards d. Translocation – part of a chromatid breaks off and goes to another chromatid
L. Binary Fission
1. The type of reproduction used by prokaryotes instead of meiosis & mitosis
2. Creates an exactly identical genetic copy
3. Creates genetic copy, then undergoes cytokinesis
M. Eukaryotic cell cycle regulation
1. Purpose a. Monitors the timing and rate of cell division b. Allows cell to adapt to certain circumstances c. Is of interest in cancer studies because it can help us understand how cancerous cells manage to escape the usual controls.
2. Cytoplasmic signals a. One hypothesis that explains some cellular control b. Theory statement
i. Signaling molecules in the cytoplasm drive cell cycle, activating certain portions at certain times
ii. These cytoplasmic markers might be produced at the end of each phase in the cycle, combining the cytoplasmic and “metabolic pathway” theory. c. Development
i. Experiments with mammalian cells grown in culture indicated that cells might adopt the phase of another cell
ii. Two cells in different phases of the cycle (G1 & S) were fused, and the nucleus of a G1 cell immediately adopted the S cell’s actions, duplicating chromosomes.
3. Cell Cycle Control System a. A cyclically operating set of molecules in the cell that both triggers and coordinates key events in the cell cycle b. Checkpoints
i. In the cell cycle, there are checkpoints: control points where stop/go signals can regulate the cycle
ii. The “restriction point” is the most important example of such points: it is the point at which the cell decides whether or not it will divide, occurring in G1.
This point is what creates G0; when a consistent “not dividing” signal is given here, the cell will stay in G0
iii. G0 can be broken by external cues like an injury, etc. c. Cell Cycle Clock: Cyclins and Cyclin-Dependent Kinases
i. The cell cycle clock depends on Cyclins, and protein Kinases
ii. Protein kinases: enzymes that activate or inactivate other proteins by phosphorylating them. When dependent on cyclins to function, they are appreviated
Cdks. These proteins can drive the cell cycle.
iii. Cyclins: A protein that exists in a cyclically fluctuating concentration in the cell.
iv. Kinases are present in a cell at a constant concentration, but they are usually in an inactive form. Cdks are dependent on kinases to function, and thus their protein activity fluctuates with [relevant cyclins].
v. Examples of this relationship are growth factors: proteins released by cells to encourage other cells to grow and divide. d. Control Examples
i. Density-dependent inhibition: crowded cells stop dividing because adjoining cells have a growth-inhibiting signal on the membrane and when they contact other cells, growth and division halts.
ii. Anchorage dependence: To divide, cells must be attached to a substratum, or they will not divide.
Diagram 1:
Diagram 2:
Diagram 3: