Models of Sequence Evolution
advertisement

Major Application Areas of Molecular Evolution
The Role of Models
The Assumption of Basic Models
The Famous Models: JC69, K80, F81, HKY85, REV,…
Finer points: Codons, Heterogeneity, Local Dependency,
overlapping constraints, Hidden Structure Dependency, Selection,
Testing Models
Challenges
Empirical Results
Substitution Patterns
Selection Strengths
Selection beyond AA: Codon Bias, signals and RNA structure
Open Questions
Models and Reality
Reality
Model
Complex
Simpler
Not precise
Precise
Deterministic
Stochastic
Dynamic (time)
Time:
continuous/discrete
Space
continuous/discrete
• Model identification: Select model that fits reality
• Properties and reasoning is all done within models
Principle of Inference: Likelihood
Likelihood function L() – the probability of data as function of parameters: L(Q,D)
LogLikelihood Function – l(): ln(L(Q,D))
If the data is a series of independent experiments L() will become a product of
Likelihoods of each experiment, l() will become the sum of LogLikelihoods of each
experiment
ˆ (D) Q as data increases.
Consistenc y : Q
true
n!
L( x, n; p )
p x (1 p)n x
(n x )! x!
l ( x, n; p) ln(
n!
) x ln( p) (n x ) ln( 1 p)
(n x )! x!
In Likelihood analysis parameter is not viewed as a random variable.
Bayesian Inference
In Bayesian analysis parameter is viewed as a random variable.
• Prior distribution. In previous example the parameter p would have to be given
a distribution for instance uniform on [0.0-1.0].
• After observing the data a posterior distribution given the data is defined.
• Inference is different between likelihood and Bayesian analysis, but many
modeling aspects are common.
• Computionally: Bayesian analysis involves integration over parameter space,
while likelihood analysis is local analysis around maximum likelihood estimate
The Purpose of Stochastic Models.
1. Molecular Evolution is Stochastic.
2. To estimate evolutionary parameters, not observable directly:
i. Real number of events in evolutionary history.
ii. Rates of different kinds of events in evolutionary history.
iii. Strength of selection against amino acid changing nucleotide substitutions.
iv. Estimate importance of different biological factors.
3. Survive a goodness of fit test.
4. Serve these purposes as simply as possible.
Comment: knowledge might be preclude models
Central Problems: History cannot be observed, only end products.
ACGTC
ACGTC
ACGCC
ACGCC
AGGCC
AGGCC
AGGCT
AGGCT
AGGGC
AGGCT
AGGTT
AGGCT
AGGTT
AGTGC
Even if History could be observed, the underlying process couldn’t !!
Simplifying Assumptions I
Data: s1=TCGGTA,s2=TGGTT
Biological setup
Probability of Data
P P(a ) * P(a TCGGTA) P(a TGGTT)
a - unknown
a
TCGGTA
TGGTT
1) Only substitutions.
s1
s2
TCGGTA
TGGT-T
s1
s2
TCGGA
TGGTT
P P(a ) * P(a TCGGA) P(a TGGTT)
a
2) Processes in different positions of the molecule are independent, so the probability for
the whole alignment will be the product of the probabilities of the individual patterns.
a
a1 a2 3
G A
G
T C
a4 a5
5
P Pi (ai ) * Pi (ai s1i ) Pi (ai s1i )
T
G
G
T
T
i 1 a
Simplifying Assumptions II
3) The evolutionary process is the same in all positions
5
P P(ai ) * P(ai s1i ) P(ai s2i )
i 1 a
4) Time reversibility: Virtually all models of sequence evolution are time reversible. I.e.
πi Pi,j(t) = πj Pj,i(t), where πi is the stationary distribution of i and Pt(i->j) the probability
that state i has changed into state j after t time. This implies that
P(a) * P
a,N1
(l1) * Pa,N1 (l1 ) = PN1 * PN1,N 2 (l1 l2 )
a
a
l1
N1
l2
=
N1
l2+l1
N2
N2
5
P P( s1i ) P( s1i s2i )
i 1
Simplifying assumptions III
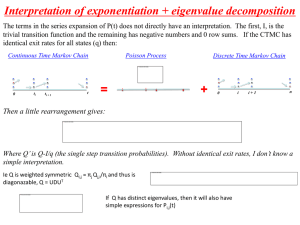
5) The nucleotide at any position evolves following a continuous time Markov Chain.
Pi,j(t) continuous time markov chain on the state space {A,C,G,T}.
lim e 0
e
qij
lim e 0
t1
e
A
Pi , j (e )
Pi ,i (e ) 1
e
qii
t2
C
C
Q - rate matrix:
T
A
F
R
O
M
A
C
G
T
C
-(qA,C+qA,G+qA,T)
qA,C
qC,A
-(qC,A+qC,G+qC,T)
qG,A
qG,C
qT,A
qT,C
O
G
qA,G
qC, G
-(qG,A+qG,C+qG,T)
qT,G
T
qA,T
qC ,T
qG,T
-(qT,A+qT,C+qT,G)
6) The rate matrix, Q, for the continuous time Markov Chain is the same at all times (and
often all positions). However, it is possible to let the rate of events, ri, vary from site to
site, then the term for passed time, t, will be substituted by ri*t.
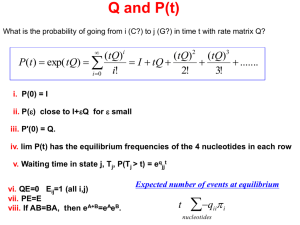
Q and P(t)
What is the probability of going from i (C?) to j (G?) in time t with rate matrix Q?
(tQ)i
(tQ)2 (tQ)3
P(t ) exp( tQ)
I tQ
.......
i!
2!
3!
i 0
i. P(0) = I
ii. P(e) close to I+eQ for e small
iii. P'(0) = Q.
iv. lim P(t) has the equilibrium frequencies of the 4 nucleotides in each row
v. Waiting time in state j, Tj, P(Tj > t) = e -(qjjt)
vi. QE=0 Eij=1 (all i,j)
vii. PE=E
viii. If AB=BA, then eA+B=eAeB.
Jukes-Cantor 69: Total Symmetry
Rate-matrix, R:
T
A
F
R
O
M
C
O
G
T
3*a
a
a
a
a
3*a
a
a
aa3*aa
aaa3*a
A
C
G
T
Transition prob. after time t, a = a*t:
P(equal) = ¼(1 + 3e-4*a ) ~ 1 - 3a
P(diff.) = ¼(1 - 3e-4*a ) ~ 3a
Stationary Distribution: (1,1,1,1)/4.
5
1
P P( s1) P( s1i s2i ) ( )5 P(T T)P(C G)P(G G)P(G T)P(A T)
i 1
4
1 1
( )5 ( )5 (1 3e 4 a )2 (1 e 4 a )3
4 4
3a
a
a
a
From Q to P for Jukes-Cantor
a
a
a
1
1
3 1
1 3 1
3a
a
a
1
a
a
3a
a
1 3 1
1
a
a
3a
1
1 3
1
3a a
a 3a
a a
i 0
a
a
3 1
1 3
1/4I
1 1
1 1
1
1
1
1
3 1
3 1
1 3 1
1 3 1
1
1
i
1
4
1 3 1
1 3 1
1
1
1
1 3
1
1 3
1
1
i
3 1 1 1
a
a
a
a
1
3
1
1
/i!
/i! I 1/4 (4at) i
1 1 3 1
3a a
i1
a 3a
1 1 1 3
1 1
1 1 4 at
e
3 1
1 3
Kimura 2-parameter model
TO
A
C
G
T
F A
-2*babab
R C
b2*baba
Q: O
G
M T
a = a*t
ab2*bab
bab2*ba
b = b*t
start
.25(1 e4b 2e2( a b) )
.25(1 e4b )
P(t):
.25(1 e4b 2e2( a b ) )
.25(1 e4b )
Felsenstein81 & Hasegawa, Kishino & Yano 85
Unequal base composition:
Qi,j = C*πj
(Felsenstein, 1981)
i unequal j
Transition/transversion & compostion bias (Hasegawa, Kishino &
Yano, 1985)
(a/b)*C*πj
Qi,j =
C*πj
i- >j a transition
i- >j a transversion
General Reversible Model
TO
A
C
F A
-
R C
G
T
agC
bgG
cgT
agA
-
dgG
egT
O G
bgA
dgC
-
fgT
M T
cgA
egC
fgG
-
• Biased symmetric.
• 6 parameters for the upper triangular matrix
• 3 parameters for the nucleotide bias
Basic Dinucleotide model: AB --> CD
From singlet models to doublet models:
Contagious Dependence:
Independence
Independence with CG avoidance
Strand symmetry
Only single events
Single events with simple double events
The Data:
100 kb non-coding
from chromosomes
22 and 10 from
mouse and human.
From Lunter & Hein,2004
Pedersen and Jensen, 2001
Siepel and Haussler, 2003
Rate variation between sites:iid each site
The rate at each position is drawn independently from a distribution, typically a G (or
lognormal) distribution. G(a,b) has density xb-1*e-ax/G(b) , where a is called scale
parameter and b form parameter.
Let L(pi,Q,t) be the likelihood for observing the i'th pattern, t all time lengths, Q the
parameters describing the process parameters and f (ri) the continuous distribution of
rate(s). Then L
L( p , Q, r ) f ( r )dr
i
i
i
i
Fast/Slowly Evolving States
Felsenstein & Churchill, 1996
positions
1
1
n
k
slow - rs
fast - rf
HMM:
• pr - equilibrium distribution of hidden states (rates) at first position
•pi,j - transition probabilities between hidden states
•L(j,r) - likelihood for j’th column given rate r.
•L(j,r) - likelihood for first j columns given j’th column has rate r.
Likelihood Recursions:
L(j,f) (L(j-1,f) p f , f L(j-1,s) ps, f )L( j, f ) L(j,s) (L(j-1,f) p f ,s L(j-1,s) ps,s )L( j,s)
Likelihood Initialisations:
L(1,f) p f L(1, f )
L(1,s) p sL(1,s)
Dayhoffs empirical approach
(1970)
Take a set of closely related
proteins, count all differences
and make symmetric difference
matrix, since time direction
cannot be observed.
If qij=qji, then
equilibrium frequencies,
pi, are all the same.
The transformation qij -->
piqij/pj, then equilibrium
frequencies will be pi.
Codon based Models
Goldman,Yang + Muse,Gaut
i. Codons as the basic unit.
ii. A codon based matrix would have (61*61)-61 (= 3661) off-diagonal entries.
i. Bias in nucleotide usage.
ii. Bias in codon usage.
iii. Bias in amino acid usage.
iv. Synonymous/non-synonymous distinction.
v. Amino acid distance.
vi. Transition/transversion bias.
codon i and codon j differing by one nucleotide (otherwise 0.0), then
apj exp(-di,j/V)
qi,j =
bpj exp(-di,j/V)
differs by transition
differs by transversion.
-di,j is a physico-chemical difference between amino acid i and amino acid j. V
is a factor that reflects the variability of the gene involved.
Measuring Selection
ThrSer
ACGTCA
ThrPro
ACGCCA
Certain events have functional
consequences and will be selected
out. The strength and localization of
this selection is of great interest.
-
ThrSer
ACGCCG
ArgSer
AGGCCG
The selection criteria could in
principle be anything, but the
selection against amino acid changes
is without comparison the most
important
ThrSer
ACTCTG
AlaSer
GCTCTG
AlaSer
GCACTG
The Genetic Code
3 classes of sites:
4
2-2
1-1-1-1
i.
4 (3rd)
Problems:
1-1-1-1 (3rd)
ii. TA (2nd)
i. Not all fit into those categories.
ii. Change in on site can change the status of another.
Possible events if the genetic code
remade from Li,1997
Possible number of substitutions: 61 (codons)*3 (positions)*3 (alternative nucleotides).
Substitutions
Number
Percent
Total in all codons
549
100
Synonymous
134
25
415
75
Missense
392
71
Nonsense
23
4
Nonsynonymous
Synonyous (silent) & Non-synonymous (replacement) substitutions
Ser
TCA
***
GGG
Ser
Thr
ACT
*
ACA
Thr
Glu
GAG
*
GGG
Gly
Met
ATG
*
ATA
Ile
Cys
TGT
*
TAT
Tyr
Leu
TTA
*
CTA
Leu
Met Gly Thr
ATG GGG ACG
* **
ATG GGT AGC
Met Gly Ser
Ks : Number of Silent Events in Common History
Ka : Number of Replacement Events in Common History
Ns : Silent positions
Na : replacement positions.
Rates per pos: ((Ks/Ns)/2T)
Example: Ks =100 Ns = 300 T=108 years
Silent rate (100/300)/2*108 = 1.66 * 10-9 /year/pos.
Thr
ACC
*
Thr
Miyata: use most silent path for calculations.
ACG
Ser
*
Arg
AGG
*
AGC
Kimura’s 2 parameter model & Li’s Model.
Probabilities:
Rates:
start
b
.25(1 e4b 2e2( a b) )
b
a
b
.25(1 e4b )
a
.25(1 e4b 2e2( a b ) )
.25(1 e4b )
Selection on the 3 kinds of sites (a,b)(?,?)
1-1-1-1
(f*a,f*b)
2-2
(a,f*b)
4
(a, b)
alpha-globin from rabbit and mouse.
Ser
TCA
*
TCG
Ser
Sites
1-1-1-1
2-2
4
Thr
ACT
*
ACA
Thr
Glu
GAG
*
GGG
Gly
Total
274
77
78
Z(at,bt) = .50[1+exp(-2at) - 2exp(-t(a+b)]
Y(at,bt) = .25[1-exp(-2bt )]
X(at,bt) = .25[1+exp(-2at) + 2exp(-t(ab)]
Met
ATG
*
ATA
Ile
Cys
TGT
*
TAT
Tyr
Leu
TTA
*
CTA
Leu
Met Gly Gly
ATG GGG GGA
* **
ATG GGT ATA
Met Gly Ile
Conserved
246 (.8978)
51 (.6623)
47 (.6026)
Transitions
12(.0438)
21(.2727)
16(.2051)
Transversions
16(.0584)
5(.0649)
15(.1923)
transition
transversion
identity
L(observations,a,b,f)=
C(429,274,77,78)* {X(a*f,b*f)246*Y(a*f,b*f)12*Z(a*f,b*f)16}* {X(a,b*f)51*Y(a,b*f)21*Z(a,b*f)5}*{X(a,b)47*Y(a,b)16*Z(a,b)15}
where a = at and b = bt.
Estimated Parameters:
1-1-1-1
2-2
4
a = 0.3003 b = 0.1871
Transitions
a*f = 0.0500
a
= 0.3004
a
= 0.3004
2*b = 0.3742 (a + 2*b) = 0.6745 f = 0.1663
Transversions
2*b*f = 0.0622
2*b*f = 0.0622
2*b
= 0.3741
Expected number of:
replacement substitutions 35.49
synonymous
Replacement sites : 246 + (0.3742/0.6744)*77 = 314.72
Silent sites
: 429 - 314.72
= 114.28
Ks = .6644 Ka = .1127
75.93
Extension to Overlapping Regions
Hein & Stoevlbaek, 95
1st
1-1-1-1
2-2
4
1-1-1-1 sites
(f1f2a, f1f2b)
(f2a, f1f2b)
(f2a, f2b)
2-2
(f1a, f1f2b)
(f2a, f1f2b)
(a, f2b)
4
(f1a, f1b)
(a, f1b)
(a, b)
2nd
pol
gag
Example: Gag & Pol from HIV
Pol
Gag
1-1-1-1
2-2
4
1-1-1-1 sites
64
31
34
2-2
40
7
0
4
27
2
0
MLE:
a=.084
b= .024
a+2b=.133
fgag=.403
fpol=.229
Ziheng Yang has an alternative model to this, were sites are lumped into the same category if they have the same configuration of positions and reading frames.
HIV1 Analysis
Hasegawa, Kisino & Yano Subsitution Model Parameters:
a*t
0.350
0.015
β*t
0.105
0.005
pA
0.361
0.004
pC
0.181
0.003
pG
0.236
0.003
Selection Factors
GAG
POL
VIF
VPR
TAT
REV
VPU
ENV
NEF
0.385
0.220
0.407
0.494
1.229
0.596
0.902
0.889
0.928
(s.d.
(s.d.
(s.d.
(s.d.
(s.d.
(s.d.
(s.d.
(s.d.
(s.d.
0.030)
0.017)
0.035)
0.044)
0.104)
0.052)
0.079)
0.051)
0.073)
Estimated Distance per Site: 0.194
pT
0.222
Statistical Test of Models
Data: 3 sequences of length L
ACGTTGCAA ...
AGCTTTTGA ...
TCGTTTCGA ...
(Goldman,1990)
A. Likelihood (free multinominal model 63 free parameters)
L1 = pAAA#AAA*...pAAC#AAC*...*pTTT#TTT where pN1N2N3 = #(N1N2N3)/L
B. Jukes-Cantor and unknown branch lengths
ACGTTGCAA ...
l1
l2
l3
TCGTTTCGA ...
L2 = pAAA(l1',l2',l3') #AAA*...*pTTT(l1',l2',l3') #TTT
AGCTTTTGA ...
Test statistics: I. (expected-observed)2/expected or II: -2 lnQ = 2(lnL1 - lnL2)
JC69 Jukes-Cantor: 3 parameters => c2 60 d.of freedom
Problems: i. To few observations pr. pattern.
Parametric bootstrap:
i. Maximum likelihood to estimate the parameters.
iii. Make simulated distribution of -2 lnQ.
ii. Many competing hypothesis.
ii. Simulate with estimated model.
iv. Where is real -2 lnQ in this distribution?
History of Phylogenetic Methods & Stochastic Models
1958 Sokal and Michener publishes UGPMA method for making distrance trees with a clock.
1964 Parsimony principle defined, but not advocated by Edwards and Cavalli-Sforza.
1962-65 Zuckerkandl and Pauling introduces the notion of a Molecular Clock.
1967 First large molecular phylogenies by Fitch and Margoliash.
1969 Heuristic method used by Dayhoff to make trees and reconstruct ancetral sequences.
1969 Jukes-Cantor proposes simple model for amino acid evolution.
1970: Neyman analyzes three sequence stochastic model with Jukes-Cantor substitution.
1971-73 Fitch, Hartigan & Sankoff independently comes up with same algorithm reconstructing
parsimony ancetral sequences.
1973 Sankoff treats alignment and phylogenies as on general problem – phylogenetic alignment.
1979 Cavender and Felsenstein independently comes up with same evolutionary model where
parsimony is inconsistent. Later called the “Felsenstein Zone”.
1979: Kimura introduces transition/transversion bias in nucleotide model in response to
pbulication of mitochondria sequences.
1981: Felsenstein Maximum Likelihood Model & Program DNAML (i programpakken PHYLIP).
Simple nucleotide model with equilibrium bias.
1981 Parsimony tree problem is shown to be NP-Complete.
1985: Felsenstein introduces bootstrapping as confidence interval on phylogenies.
1985: Hasegawa, Kishino and Yano combines transition/transversion bias with unequal equilibrium
frequencies.
1986 Bandelt and Dress introduces split decomposition as a generalization of trees.
1985-: Many authors (Sawyer, Hein, Stephens, M.Smith) tries to address the problem of
recombinations in phylogenies.
1991 Gillespie’s book proposes “lumpy” evolution.
1994 Goldman & Yang + Muse & Gaut introduces codon based models
1997-9 Thorne et al., Sanderson & Huelsenbeck introduces the Almost Clock.
2000 Rambaut (and others) makes methods that can find trees with non-contemporaneous leaves.
2000 Complex Context Dependent Models by Jensen & Pedersen. Dinucleotide and overlapping
reading frames.
2001- Major rise in the interest in phylogenetic statistical alignment
2001- Comparative genomics underlines the functional importance of molecular evolution.
References: Books & Journals
Joseph Felsenstein "Inferring Phylogenies” 660 pages Sinauer 2003
conceptual issues.
Excellent – focus on methods and
Masatoshi Nei, Sudhir Kumar “Molecular Evolution and Phylogenetics” 336 pages Oxford University Press Inc,
USA 2000
R.D.M. Page, E. Holmes “Molecular Evolution: A Phylogenetic Approach”
352 pages 1998 Blackwell Science (UK)
Dan Graur, Li Wen-Hsiung “Fundamentals of Molecular Evolution” Sinauer Associates Incorporated 439 pages
1999
Margulis, L and K.V. Schwartz (1998) “Five Kingdoms” 500 pages Freeman
A grand illustrated tour of the tree of life
Semple, C and M. Steel “Phylogenetics” 2002 230 pages Oxford University Press
Very mathematical
Yang, Z. (2006) Computational Molecular Evolution OUP
Journals
Journal of Molecular Evolution :
http://www.nslij-genetics.org/j/jme.html
Molecular Biology and Evolution : http://mbe.oupjournals.org/
Molecular Phylogenetics and Evolution : http://www.elsevier.com/locate/issn/1055-7903
Systematic Biology - http://systbiol.org/
J. of Classification -
http://www.pitt.edu/~csna/joc.html
References: www-pages
Tree of Life on the WWW
http://tolweb.org/tree/phylogeny.html
http://www.treebase.org/treebase/
Software
http://evolution.genetics.washington.edu/phylip.html
http://paup.csit.fsu.edu/
http://morphbank.ebc.uu.se/mrbayes/
http://evolve.zoo.ox.ac.uk/beast/
http://abacus.gene.ucl.ac.uk/software/paml.html
Data & Genome Centres
http://www.ncbi.nih.gov/Entrez/
http://www.sanger.ac.uk