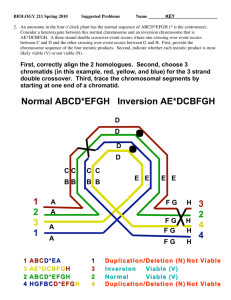
GA Intro [1]
advertisement
![GA Intro [1]](http://s3.studylib.net/store/data/009560395_1-ec7b77c71331affecb797c1c6c2db86f-768x994.png)
Crossovers and
Mutation
Richard P. Simpson
Genotype vs. Phenotype
The genotype of a chromosome is just the basic data
structure (it bits in a binary chromosome)
The Phenotype is the object that the chromosome
represents.
For example the bit pattern 10110111011101
could be used to represent a particular polygon such as
The polygon is the phenotype and the binary pattern is
the genotype. (think in terms of yourself).
Crossover and Mutations
10110111011101
mutate
101010111011011
1 Point Binary Crossover
Choose a random point on the two parents
Split parents at this crossover point
Create children by exchanging tails
Pc typically in range (0.6, 0.9)
Performance with 1 point binary
crossover
more likely to keep together genes that are
near each other
Can never keep together genes from opposite
ends of string
This is known as Positional Bias
Can be exploited if we know about the
structure of our problem, but this is not usually
the case
n-point binary crossover
Choose n random crossover points
Split along those points
Glue parts, alternating between parents
Generalisation of 1 point (still some positional
bias)
Uniform Binary Crossover
Assign 'heads' to one parent, 'tails' to the
other
Flip a coin for each gene of the first child
Make an inverse copy of the gene for the
second child
Inheritance is independent of position
Binary Mutation
Alter each gene independently with a probability pm
pm is called the mutation rate
Typically a small number between 1/pop_size and 1/
chromosome_length.
Crossover OR mutation?
What effect does each of these have on the
evolution process.
See paper Crossover or Mutation by William
M. Spears
See Genetic Algorithms:The crossovermutation Debate by Nuwan Senaratna, 2005
GA operator roles: disruption and
construction.
Generally mutation causes more disruption
and crossover causes more construction.
Disruption vs. Construction on a
landscape
Disruption (exploration) is
individual
Construction (exploitation)
climbs the hills!
Of course what actually happens
depends on the problem and its
method of chromosome coding.
IE Use both.
Crossover or Mutation
Only crossover can combine information from two
parents
Only mutation can introduce new information (alleles)
Crossover does not change the allele frequencies of
the population (thought experiment: 50% 0’s on first
bit in the population, ?% after performing n
crossovers)
To hit the optimum you often need a ‘lucky’ mutation
Representations other than binary
Gray coding of integers (still binary chromosomes)
Gray coding is a mapping that means that small changes in
the genotype cause small changes in the phenotype (unlike
binary coding). “Smoother” genotype-phenotype mapping
makes life easier for the GA
Nowadays it is generally accepted that it is better to
encode numerical variables directly as
Integers
Floating point variables
Gray Codes
When you count up or down in binary, the
number of bit that change with each digit
change varies.
From 0 to 1 just have a single but
From 1 to 2 have 2 bits, a 1 to 0 transition and
a 0 to 1 transition
From 7 to 8 have 3 bits changing back to 0
and 1 bit changing to a 1
For some applications multiple bit changes
cause significant problems.
Gray Code
Contrast of bit changes
Val
0
1
2
3
4
5
6
7
0
Bin
000
001
010
011
100
101
110
111
000
Chg
1
2
1
3
1
2
1
3
Gray
000
001
011
010
110
111
101
100
000
Chg
1
1
1
1
1
1
1
1
Integer representations
Some problems naturally have integer variables, e.g.
image processing parameters
Others take categorical values from a fixed set e.g.
{blue, green, yellow, pink}
N-point / uniform crossover operators work
Extend bit-flipping mutation to make
“creep” i.e. more likely to move to similar value
Random choice (esp. categorical variables)
For ordinal problems, it is hard to know correct range for
creep, so often use two mutation operators in tandem
Integer example
There is a well known class of math problems
called Diophantine equations. These are
multivariate equations where the solutions
are require to be integer. For example
Assume we want three integers x,y,z such
that 0 = 2x – 3y2 – 4z OR we get as close as
we can to zero! In a GA that searches for
these guys we have chromosomes that look
like
X
Y
Z
0 = 2x – 3y2 – 4z
Possible Fitness function
Fitness(x,y,z)=|2x – 3y2 – 4z|
Possible Crossover
one point crossover, between variables
Possible Mutator using random deviate d.
pick a random gene X,Y or Z and change it
with low probability. Maybe vary it slightly
X = X +/- d for some d ?!
What happens if d is small? Large?
Real Coded Genetic Algorithms
In this type of GA a chromosome is usually an
array of floating point values. It is often
applied to optimize some real valued function.
Mutations are probabilistically performed on a
real value by adding a random deviate in
some range from –d to +d. One can select
this deviation from a Gaussian (normal)
distribution or from a uniform distribution (see
rand()).
Normal Distribution about 0
http://www.cplusplus.com/reference/random/normal_distribution/
De Jong’s classical test functions
A lot of test functions can be found here
http://www.sfu.ca/~ssurjano/optimization.html
Real Crossovers
There are a lot of options here
The obvious n point crossovers where each allele
value in offspring z comes from one of its parents
(x,y) with equal probability
Intermediate (a blending scheme)
exploits idea of creating children “between” parents (hence
a.k.a. arithmetic recombination)
zi = xi + (1 - ) yi where : 0 1.
The parameter can be:
• constant: uniform arithmetical crossover
• variable (e.g. depend on the age of the population)
• picked at random every time
Do it for a single gene
• Parents: x1,…,xn and y1,…,yn
• Pick a single gene (k) at random, (or more!)
• child1 is:
x1 , ..., xk , yk (1 ) xk , ..., xn
• reverse for other child. e.g. with = 0.5
Simple arithmetic crossover
• Parents: x1,…,xn and y1,…,yn
• Pick random gene (k) after this point mix
values. The child1 is:
x , ..., x , y
(1 ) x
1
k
k 1
k 1
, ..., y (1 ) x
n
n
• reverse for other child. e.g. with = 0.5
What if =0 ?
Permutations Chromosomes
Ordering/sequencing problems form a special type
Task is (or can be solved by) arranging some
objects in a certain order
Example: sort algorithm: important thing is which elements
occur before others (order)
Example: Travelling Salesman Problem (TSP) : important
thing is which elements occur next to each other
(adjacency)
These problems are generally expressed as a
permutation:
if there are n variables then the representation is as a list of
n integers, each of which occurs exactly once
Permutations
Mutations
if every chromosome is a permutation of a set,
say {1,2,…,n} then a mutation must generate
another mutation.
The mutation operator must be applied to the
entire genome.
Two simple mutation schemes
Pick two allele values at random
Move the second to follow the first, shifting
the rest along to accommodate
Note that this preserves most of the order and
the adjacency information
Pick two alleles at random and swap their
positions
Preserves most of adjacency information (4
links broken), disrupts order more
Inversion and Scramble Mutations
(inversion) Pick two alleles at random and
then invert the substring between them.
Preserves most adjacency information (only
breaks two links) but disruptive of order
information
(scramble) Pick a subset of genes at random
Randomly rearrange the alleles in those
positions
Crossover operators for Perms
Partially mapped crossover (PMX)
Order crossover (X)
Uniform order crossover
Edge recombination
There are many other that we will not discuss.
Partially Mapped Crossover
Given two parents s and t, PMX randomly
picks two crossover points. The child is
constructed in the following way. Starting
with a copy of s, the positions between the
crossover points are, one by one, set to the
values of t in these positions. This is performs
by applying a swap to s. The swap is defined
by the corresponding values in s and t within
the selected region.
PMX example
No
change
6 2 3 4 1 7 5
6 2 3 1 4 7 5
6 2 4 1 3 7 5
6 2 4
5
3 1
4 3
1 7 6
5
6 2 4
5
3 1
4 3
1 7 6
5
6 2 4
5
3 1
4 3
1 7 6
5
First offspring
6 2 4
3 1
4 3
1 7 5
For the second offspring just swap the parents
and apply the same operation
Order Crossover
This crossover first determines to crossover
points. It then copies the segment between
them from one of the parents into the child.
The remaining alleles are copied into the
child (l to r) in the order that they occur in the
other parents.
Switching the roles of the parents will
generate the other child.
Order Crossover Example
1 2 3 4 5 6 7 8 9
4 5 6 7
3 4 7 2 8 9 1 6 5
The remaining alleles are 1 2 3 8 9.
Their order in the other parent is 3 2 8 9 1
3 4 7 2 8 9 1 6 5
3 2 8 4 5 6 7 9 1
Uniform Order Crossover
Here a randomly-generated binary mask is
used to define the elements to be taken from
that parent.
The only difference between this and order
crossover is that these elements in order
crossover are contiguous.
1 2 3 4 5 6 7 8 9
1 1 0 1 0 0 0 1 0
1 2 3 4 7 9 6 8 5
3 4 7 2 8 9 1 6 5
offspring
Edge Recombination
This operator was specially designed for the
TSP problem.
This scheme ensures that every edge (link) in
the child was shared with one or other of its
parents.
This has been shown to be very effective in
TSP applications.
Constructs an edge map, which for each site
lists the edges available to it from the two
parents that involve that city. Mark edges that
occur in both with a +.
Inversion Transformations
This scheme will allow normal crossover and
mutation to operate as usual.
In order to accomplish this we map the
permutation space to a set of contiguous
vectors .
Given a permutation of the set {1,2,3,…,N} let
aj denote the number of integers in the
permutation which precede j but are greater
than j. The sequence a1,a2,a3,…,an is called
the inversion sequence of the permutation.
The inversion sequence of 6 2 3 4 1 7 6 is
4111200
There are 4 integers greater than 1
Inversion of Permutations
The inversion sequence of a permutation is
unique! Hence there is a 1-1 correspondence
between permutations and their inversion
sequence. Also the right most inv number is
y
0 so dropped.
1
12
123
(0 0)
21
1
132
312
321
231
213
(0 1)
(1 1)
(2 1)
(2 0)
(1 0)
0
x
0
1
2
Inversions Continued
What does a 4 digit permutation map to?
1234 -> (0 0 0)
2134 -> (1 0 0)
4321 -> (3 2 1)
2413 -> (2 0 1)
1423 -> (0 1 1)
etc
Maps to a partial 3D lattice structure
Converting Perm to Inv
Input perm: array of permutation
Output: inv: array holding inv sequence
For (i=1;i<=N;i++){
inv[i]=0;
m=1;
while(perm[m]<>i){
if (perm[m]>i )then inv[i]++;
m++;
}
Convert inv to Perm
Input: inv[]
Output: perm[]
For(i=1;i<=N;i++){
for(m=i+1;m<=N;m++)
if (pos[m]>=inv[i]+1)pos[m]++;
pos[i]=omv[i]+1;
}
For(i=1;i<=N;i++) perm[i]=i;