File - Jesse H. Ruben
advertisement

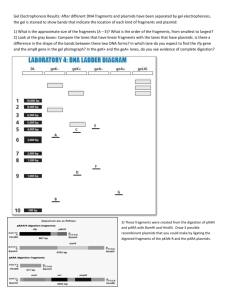
Jesse Ruben Partner Roman Verner BMB 442 Recombinants and Transformation Introduction The goal of this experiment was to take two antibiotic resistance genes for ampicillin and kanamycin from plasmids pAMP and pKan and recombining them together. Then by transforming this new plasmid into E. coli, the E. coli will then become resistant to these two antibiotics. We then can calculate the efficiency of this experiment and percentage of successful pKan plasmids made in molecules. Restriction Enzymes are used in order to break the phospodiester bonds around the different antibiotic resistant gene on the two original plasmids and a gel electrophorisis was made to make sure the DNA was broken. The broken DNA has a stick end that is recombined at compatible ends of other broken fragments by a DNA ligase and another gel was made to check the success of this process. This new multi-resistant plasmid is transformed, pulled into the cell that is known not to have either gene and uses the foreign plasmid, which now can express the resistance from transformed plasmid. Cells that can transform a plasmid are normally are called competent, the ability to absorb outside plasmids, but not all cells can do this. E.coli does not have competence, therefore a procedure is done so that the membrane of the E. coli will be artificially competent-like, and absorb this multiresistant plasmid. By plating the E. coli on LB-plates that contain the two antibiotics, colonies which grew shows how many cells transformed this plasmid. Due to the high unpredictable chances that the breakage and recombination of the DNA wasn't 100% accurate and other plasmids were Page 1 of 12 made, or remained as pAMP and pKan, plates were used that contained only one antibiotic and the colonies were then counted on these plates as well. These plating proved that our procedure rendered the E. coli to become competent and transform the different plasmids. The counted colonies are then used to find the percentage of successful transformation. The first part of the lab required us to use two 1.7 ml mircofuge tubes to set up the digestion for plasmids, pAMP and pKan, which contain the two desired antibiotic-resistant genes want for recombination. By the use of both BamHl and HindIII restriction enzymes, the two plasmids are digested and broken into two parts where these enzymes cut the backbone of specific DNA sequence on a double strain DNA. The restriction enzymes leave two sticky ends which can only be "glued" back together with a compatible sticky ends by the DNA ligase, which is used later. 6ul of Each plasmid was then put into their individual tube and 1 ul of each restriction enzyme was distributed into both tubes. Then 2.5 ul of 10x restriction buffer was the add, which provided the restriction enzymes with a proper pH and ionic conditions to maximize their activity. 14.5 ul of dd H2O was used to make each tube volume contain 25 ul. This mixture was then incubated at 37oC for 35 minutes to complete the digestion reaction. After the digestion 12 ul of our each digestion were separated and given 3 ul of 5x Sample buffer(total 15 ul) and for a 1.0% gel electrophoresis. The 15ul of each reaction was load onto the gel along with a 10 ul of a DNA ladder for each partner. The last two wells were also loaded with 10ul of the untreated pAMP and pKAN plasmid for comparison, to see if our digestions worked. The gel was then photographed and the leftover of each digestion was then incubated to inactivate the restriction enzymes, so the ligation process would work without being cut again. Page 2 of 12 The ligation started by taking 7.5 ul of each digested pAmp and pKan into the one ligation tube, along with 1 ul of DNA ligase and 3 ul of 10x ligase buffer, this provides proper conditions to activate the ligase enzymes (total 19 ul). The mixture is then levels to 30 ul with 11ul of dd H2O. This ligation mixture was then left in an ice bucket, which then is used for the transformation and competence manipulation. The strain HB101 E. coli is used because it's genome doesn't encode for Amp or Kan resistance. Our goal was to transform the ligated plasmid into the cell by manipulating the environment to induce E. coli to become competent, since this doesn't happen naturally for E coli, we had to created the right conditions. A prepared incubation of HB101 was given in a centrifuge tube and places in an ice bucket, by keeping the cells cold their membranes motility slows down and allows for us manipulate it. Then by centrifuging at 5,000 rpm for 5 minutes in a cold centrifuge, allowed us to remove the supernatant and take the pellet of cells and resuspend them in 10 ml of ice-cold CAST solution and placed on ice again. Due to the membranes naturally negative charge, the plasmid that we are trying to transform will only be repelled, since DNA has a negative charge as well. To fix this problem the CAST solution contains Calcium which is positively charged, this can interact with the membrane allowing the DNA to pass by. Normally the membrane is so motile that the Ca2+ has a hard time interacting with it, but since we keep the cells cold the calcium was then able to interact with the membrane. The mixture is then centrifuges again at 4000 rpm for 5 minutes and the supernatant is completely removed again. We then resuspend the pellet with 1.2 ml of ice-cold CAST and separate 0.2 ml of this mixture into four 12ml culture tubes labeled, one for each partner( 4ul Page 3 of 12 of ligation DNA), one negative control (NO DNA added) and one positive control ( 2ul untreated pKan DNA). The four tubes were then placed on ice for 20 minutes, which keeps the membranes matrix slow. We then heat shocked the cells for 1.5 minutes at 42oC, at this point the outside of the cell is warming up, but the inside of the cell is still cold. With the thermal transfer opening up the membrane pores, the DNA plasmid is that was used in each tube was (my ligation, partners ligation, pKan plasmid, No DNA) was also taken in. The cells are immediately cooled, trapping the DNA inside of it, thus manipulating the competence of the HB101 cell. The cells were then transferred into a culture tube of 0.8 ml of LB and incubated at 37oC for 45 minutes, this allowed the cells to recover by diluting the Ca2 and allowed them to express the new plasmid they just absorbed. By plating each reaction on a desired antibiotic plates, we can the count the number of colonies grown and judge the complete transformation of our ligation and which resistance was expressed. Each partner took 250ul of the transformed cells and placed them on the plate with the Amp antibiotic, another with Kan antibiotic and one with both antibiotics to see if the plasmid that was absorbed had one or both genes. 250 ul of the negative control with no DNA was placed on the LB plate with AMP and another with KAN antibiotic to test if the antibiotics even worked properly. And 100 ul of the positive control, which contained transformed pKan DNA only, was placed on the pKan LB plate and LB plate with no antibiotics to count how many cells were being plated, but first a serial dilution was made starting with 100 ul. After the final dilution, 50ul of the 106 tube was plated on the normal LB plate, which was used to count the number cells being plated and gives us the percentage of transformed efficiency. Page 4 of 12 Another gel is then made, but at 0.8%, which is used to check our ligation product and success, this gel was shared amongst 7 people and the ligation did work, which is mentioned in the results. This means that some of the cells were being transformed with the correct plasmid and would grow on the agarose plates with antibiotics. Results Gel Figure 1: After digesting the two plasmids we then did a ligation of the restriction fragments. But before assuming that the DNA had been properly digested, we checked the product with a gel Electrophoresis. Each Student placed 10 ul of a molecular weight DNA ladder their before digestion samples. Then 15 ul of each digested reaction in the next wells, my partner had placed his reactions in the 2nd and 3rd well and placed mine in the 5th and 6th well. The last two wells contained 10 ul of the uncut plasmid pAmp and pKan, 7th and 8th well respectively, which were placed in the gel for comparison for the reactions to check if a proper digestion took place. Table 1 (row A and B) are rough size estimates of the fragments were then recorded using the DNA ladder as a reference and Table 1 (row C) shows the actual base pair sizes in each of band of the well. *There were different amount of bands that appeared in the uncut lanes of pKan and pAmp, Page 5 of 12 which represented different topiosomers. The uncut pKan plasmids had two clear bands, neither of them were the linearized topiosomer since the plasmid was uncut (this would have appear as a third line between these two bands). The two bands that appeared were the supercoiled plasmid, which traveled further due to its tightly packed matrix, and the nicked topiosomer, which traveled slower due to its circular structure. The pAmp on the other hand had a different result that still contained the supercoiled and the nicked topiosomer, but also other topiosomers appeared above them. These other bands are due to plasmid multimers, which are two or more plasmids that are linked together or chained around each other, this results in a much slower distance traveled through the gel since more base pairs are attached. * The BP size of the extra bands in lane 7 are multimer plasmids and can't be estimated due to the type of gel used, which is meant for estimating lower base pair fragments Table 1 Lanes Estimated Band 1 Estimated Band 2 Actual/Expected sizes A B C Lane 1 DNA ladder 3755 bp Lane 2 Romans pAmp 2333 bp Lane 3 Romans pKan 4539 bp Lane 4 DNA ladder 1,2,3,4…., 10 Kb 4194 bp Lane 5 My pAmp Fragments 3800 bp 800 bp 3755 bp 784 bp Lane 6 My pKan Fragments 2500 bp 1700bp 2333 bp 1861 bp Lane 7 Uncut pAmp 3600 bp 10000 bp 4539 bp /////// Lane 8 Uncut pKan 3100 bp 9500 bp 4194 bp ///// The next step to analyzing the gel was to determine if both reactions were successfully broken into two fragments, since each restriction enzyme breaks one specific site then two fragments will be made. The pKan digestion was clearly broken and formed two bands different from the plasmid band, but the pAmp digestion encountered a problem in determining if the digestion was complete or not. One of the sizes of the fragment’s were 3755 base pair long and Page 6 of 12 had traveled close to the same distance of the supercoiled plasmid of pAmp, around 3800 base pair. The solution can be deduced first by determining if the plasmid was only partial digested by only one restriction enzyme then the plasmid would be linearized and travel slower than the supercoiled plasmid which would clearly produce a distinguishable band, in this experiment we did not have a partially digested plasmid. Now we must determine if neither enzyme worked and if it left a supercoil band, which produces the problem, since the fragment of pAmp traveled the same distance as the supercoil plasmid. Visually this can’t be distinguished, but by deducing the fact that there were no linearized plasmid would indicates that either both restriction enzymes work or that neither of the enzymes worked. Clearly we had the second band from the digestion, at 784bp, which means most likely the digestion did work and all of the enzymes did do their job correctly. So all of the fragments agreed with the predicted positions made from the actual size of each fragment and plasmid. After completing the first gel to make sure the digestion took place we then preformed a ligation of these our two digestions by mixing them together and using the protocol mentioned in the introduction. Before proceeding with the transformation process, my partner and I first preformed the second gel, along with partners of other groups, to check if the ligation was successful. The gel showed new large DNA plasmids, but this doesn’t tell us if we produced the desired plasmid with both genes. Gel figure 2: Page 7 of 12 lane 1 Lane 2 Lane 3 Lane 4 Lane 5 Lane 6 Lane 7 Lane 8 DNA ladder My Ligation Romans Ligation Dannys Ligation Jennys Ligation X ligations Y ligation Z ligation The plasmids traveled all along the gel and created multiple bands, which meant that the ligation had happened and different fragments connected together creating a diverse number of plasmids. The diversity was based on the number of fragments that have been glued together creating a giant circle, and/or plasmids that were looped together forming a chain. This diversity was a clear indicator that the ligation had worked, but there were some fragments that had missed the ligation, which are indicated by having less base pairs and still traveled further down the gel. As expected ligation wasn’t 100% successful in producing the desired multi-resistant plasmid we were trying to create, so we must take the ligation reaction and transform them into a cell to find out how many desired plasmids were made. If the ligation didn’t work we would expect to see the still digested fragments in gel figure 1, but in one well. After checking the success of the ligation we then manipulated the E. coli HB101 into competent state, which would absorb the ligated plasmids and express the proteins for the Page 8 of 12 resistance to either or both antibiotics. The cells grew on LB plates with an antibiotic, depending on the uptake transformed of plasmid; the cell would grow or die on the plate with one or both antibiotics. The procedure is mentioned in the introduction and Table 2 indicates the recorded number of Type of plate KAN AMP Amp/Kan LB 21 15 10 ---- The negative control was made Jesse's Ligation to eliminate any possibility of Romans's Ligation 30 12 8 ---- No DNA (Negative Control) 0 0 ---- ---- pKan (Positive Control) 261 ---- ---- 57 colonies grown on each plate. corruption in the antibiotics, so E. coli HB101 with neither resistant gene didn't grow on any antibiotic plate, which indicates that both antibiotics worked and we didn’t count any E. coli without the proper genes on a specific plate. The positive control was made so that we can calculate the efficiency of transformation and the percentage of successful plasmids made. The efficiency was calculated by the number of transformed bacteria, positive control KAN plate which was serial diluted to 106, divided on the by the total number of bacteria plated, positive control LB plate: 261 colonies of pKan E. coli were counted on the Kan-LB plate of 250ul and 57 colonies of pKan E .coli were counted on the 106 serial dilution LB plate of 100 ul. Transformation effiency = # 𝒐𝒇 𝒕𝒓𝒂𝒏𝒔𝒇𝒐𝒓𝒎𝒆𝒅 𝒃𝒂𝒄𝒕𝒆𝒓𝒊𝒂 𝑻𝒐𝒕𝒂𝒍 # 𝒐𝒇 𝒃𝒂𝒄𝒕𝒆𝒓𝒊𝒂 𝒑𝒍𝒂𝒕𝒆𝒅 e= 𝟐𝟔𝟏 𝒄𝒆𝒍𝒍𝒔 𝟏𝟒.𝟐𝟓 𝒙 𝟏𝟎𝟔𝒄𝒆𝒍𝒍 /𝒎𝒍 = .000018315 ≈ .0018% Page 9 of 12 Another way to represent the efficiency of the plasmid is to calculate the percentage of successful plasmids made. By converting the base pair of the pKan plasmid, 4194 bp, into gram per moles, 1 bp = 550 𝑔𝑟𝑚𝑎𝑠 𝑚𝑜𝑙𝑒 then by multiplying the amount of grams put into the reaction, 2ul of 5ng/ul, we can then finding out how mole of pKan were put into the transformation reaction and convert that into molecules. 4194 bp x 10ng x 550 𝑔𝑟𝑎𝑚 𝑚𝑜𝑙𝑒𝑠 𝑚𝑜𝑙𝑒𝑠 2,306,700𝑔𝑟𝑎𝑚𝑠 = 2,306,700 𝑔𝑟𝑎𝑚𝑠 𝑚𝑜𝑙𝑒 ≈ 4.335 x 10-15 moles 4.335 x10-15 moles x 6.022 x1023 molecules 𝑚𝑜𝑙𝑒𝑠 = 2,610,537,000 molecules Then by multiplying the percentage plated to the number of total molecules added from the start, we then can calculated the total number of pKan molecules in the plated transformation. 2,610,537,000 mol. x 250ul 800 𝑢𝑙 = = 815,792,812.5 Total pKan mol. plated Now by dividing the number of successful plasmids (counted on pkan plate) by the total pKan molecules plated and multiply by 100% we can then calculate the goal of this experiment, which is instead of just calculating the percentage of cell that transformed Page 10 of 12 and grew, we also calculated percentage of molecules that were transformed into the cells as well. 261 coloines 815,792,812.5 𝑚𝑜𝑙.𝑝𝑙𝑎𝑡𝑒𝑑 x 100% = 3.199 x 10-5 % of successful plasmids Discussion The counted cells on each antibiotic plate had very close numbers of cells that absorbed a plasmid with one or both of the resistance genes. The known hypothesis of Recombination and Transformation is that the origin of replication (ORC) gene sequence allows the plasmid to be functional and expressed in the cell. The fragment that contains pKan doesn't have the ORC gene, but the pAmp fragment does have the ORC gene. Therefore whichever plasmid pAmp is ligated to will also have function, unlike pKan which needs to ligate to a fragment that contains ORC, in order for KAN resistance to be expressed. According to this understanding pAMP will be expressed more often than pKan and the likelihood that pAmp and pKan fragments ligated together expressing both is a 4:2:1 ratio. In this experiment the pAmp resistance did appear more frequently than pKan, and the plasmid with both resistances happened less often as well, the frequency was a 21:15:10 ≈ 4:3:2, which are very close to hypothesized frequency. If we wanted to get a higher chance of obtaining cells with only desired plasmid, a technique of gel purification could had been done. By slicing out the desired digested fragments in the gel and melting the gel. We then could purify the fragments out of the Page 11 of 12 gel and perform a ligation only between the fragments with pKan and pAmp. The ligation reaction would have been in favor of creating only our multi-resistant plasmid and we then could transform them into the HB101 cells. The numbers calculated to get the transformation efficiency and percentage of successful plasmids tells us that the transformation of the plasmid is a very hard process to overcome on a cell that needs to be manipulated for competence. If the conditions stayed the same and more DNA was used then more cells would have transformed and the transformation efficiency and percentage of successful plasmids would Page 12 of 12