LECT31 replic
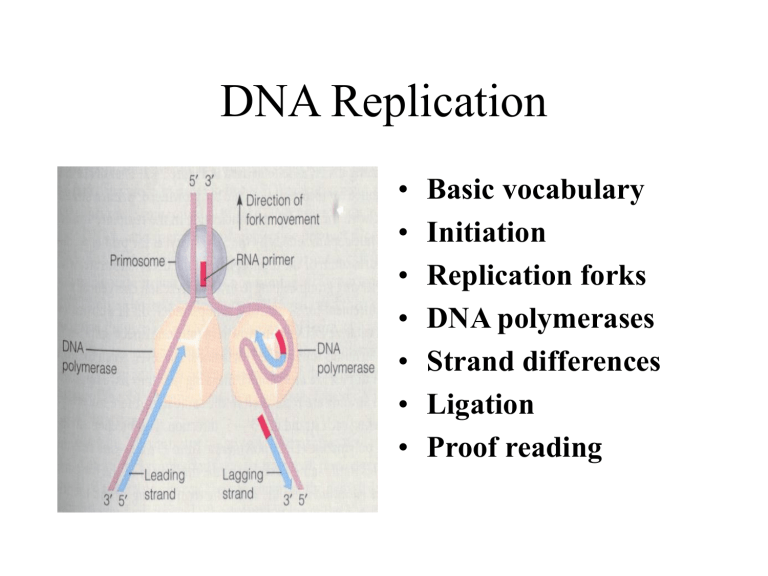
DNA Replication
• Basic vocabulary
•
Initiation
• Replication forks
•
DNA polymerases
• Strand differences
•
Ligation
• Proof reading
Replication Vocabulary
• Semiconservative vs conservative
• DNA polymerase
• Theta structures
• Replication fork
• Unidirectional vs bidirectional
• Okazaki fragments
• Template strand
• Primer
• Leading strand
• Lagging strand
• DNA ligase
• RNA primers
• Primase
• Pol 1, Pol III
• Processivity
• Nick translation
• Proofreading
• Klenow fragment
• Primosome
• DNA binding proteins
• 3’ and 5’ Exonucleases
DNA BIOSYNTHESIS
3 Foundation Studies
•
Meselson and Stahl-Conservative vs
Semiconservative replication
• Cairns’ Replicating Fork
• Okazaki’s fragments
Meselson and Stahl
Heavy
15 N
Light
14 N
0 1-3
>10
Conservative
CsCl
15 NH
4
Cl
14 N, 15 N
Hybrid band
Observed
0 1-3 >10
Semiconservative
Semiconservative
Cairns’ Replicating Fork
5’
3’
“The movement of the replicating fork is unidirectional, i.e., synthesis occurs on both strands but the fork is moving in only one direction
Kornberg “DNA synthesis addition of dNTP to the 3’ end of the growing chain of nucleotides
ERGO:
3’
5’ 3’
5’
Lagging Strand
Leading Strand:
Something is wrong
Okazaki fragments
Replication is in the direction of the moving fork
Mechanism of Chain Elongation
CH
2
O
B
1
3’-OH
H O H
O O O
O-P-O-P-OP-OCH
2
O
O O O
CH
2
O
B
1
B
2
O
P
O
O
O
H
CH
2
O
B
2
H O H
H O H
SUMMARY OF DNA SYNTHESIS dATP, dGTP, dCTP, dTTP + Template + Primer
5’
RNA Primer
3’ OH
U A G G
A T C C G A T G A C T T
3’
3’ end
5’
Template
DNA synthesis involves addition of dNTP
Template
Stages and Events in Replication
Initiation:
Proteins recognize the origin
Parental strands separate
Primosome initiates synthesis at replication fork
Elongation:
Parental strands unwind at the forks
Replisome complex moves along DNA
Leading strand is replicated continuously
Lagging stand is replicated discontinuously
Termination:
Duplicate chromosomes separate
Replication Fork
Directionality
Loop
Replication must always proceed in a 5’ to 3’ direction
What is a Primosome?
Ans: A 600 kD protein assembly composed of at least 7 individual proteins that take part in replication
What function does it perform?
Ans: Primosomes conduct the initial phases of replication,
They unwind the DNA, separate and keep the strands apart, lay down an RNA primer
Why is it needed?
Ans: Because DNA polymerase cannot initiate the synthesis of a DNA chain, and leading and lagging strands must have a 3’OH to start off and extend
Major Primosome Composition
Primosomes assemble on single-stranded DNA in a unique region called the primosome assembly site (pas)
One of the primosomes 7 proteins displaces SSB in order to localize the primosome.
PriA
DnaB
Primase
ATP driven, displaces SSB (single strand binding protein) unwinds priming
An RNA polymerase (DnaG)
DNA Polymerases
Polymerase I (low processivity)
5’-3’ exonuclease to remove RNA primer
Lagging strand fill in
3’-5’ exonuclease
Polymerase II
DNA repair
Polymerase III (high processivity)
Leading and lagging strand DNA synthesis
3’-5’ exonuclease
Polymerase III
•
Mwt = 130,000
•
10 different polypeptide chains
•
Core polymerase =
,
, and
subunits
•
2 molecules of
cause dimerization
• and
allow pol III to bind
subunit
• subunit forms sliding clamp that is responsible for high processivity
• Assembled protein is called “holoenzyme”
DNA
Beta Subunit Dimer is responsible for high processivity of DNA Pol III
Beta Subunit as a dimer forms a ring to clamp onto the DNA
Processivity >5000
3’ 5’
5’ 3’
Pol I, III
Template strand
Proof Reading
New strand
(clips from 3’ end)
Pol I only
Absent in Klenow fragment
Proofreading
Template
3’ end
3’exonuclease
Bound dCTP