3. G-protein-coupled receptors
advertisement

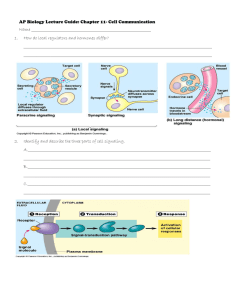
Patrick An Introduction to Medicinal Chemistry 3/e Chapter 6 PROTEINS AS DRUG TARGETS: RECEPTOR STRUCTURE & SIGNAL TRANSDUCTION Part 2: Sections 6.3 - 6.6 ©1 Contents Part 2: Sections 6.3 - 6.6 3. G-protein-coupled receptors (7-TM receptors) 3.1. Structure - Single protein with 7 transmembrane regions 3.2. Ligands 3.3. Ligand binding site - varies depending on receptor type 3.4. Bacteriorhodopsin & rhodopsin family (2 slides) 3.5. Receptor types and subtypes (2 slides) 3.6. Signal transduction pathway a) Interaction of receptor with Gs-protein (3 slides) b) Interaction of s with adenylate cyclase (2 slides) c) Interaction of cyclic AMP with protein kinase A (PKA) (4 slides) 3.7. Glycogen metabolism - triggered by adrenaline in liver cells (2 slides) 3.8. GI proteins 3.9. Phosphorylation 3.10. Drugs interacting with cyclic AMP signal transduction 3.11.Signal transduction involving phospholipase C (PLC) (2 slides) 3.12. Action of diacylglycerol (2 slides) 3.13. Action of inositol triphosphate (2 slides) 3.14. Resynthesis of PIP2 [29 slides] ©1 3. G-protein-coupled receptors (7-TM receptors) 3.1 Structure - Single protein with 7 transmembrane regions Extracellular loops NH2 N -Terminal chain Membrane VII VI V IV III II I Transmembrane helix G-Protein binding region HO2C C -Terminal chain Variable intracellular loop Intracellular loops ©1 3. G-protein-coupled receptors (7-TM receptors) 3.2 Ligands • Monoamines e.g. dopamine, histamine, noradrenaline, acetylcholine (muscarinic) • Nucleotides • Lipids • Hormones • Glutamate • Ca++ ©1 3. G-protein-coupled receptors (7-TM receptors) 3.3 Ligand binding site - varies depending on receptor type Ligand A B C D A) Monoamines - pocket in TM helices B) Peptide hormones - top of TM helices + extracellular loops + N-terminal chain C) Hormones - extracellular loops + N-terminal chain D) Glutamate - N-terminal chain ©1 3. G-protein-coupled receptors (7-TM receptors) 3.4 Bacteriorhodopsin & rhodopsin family • Rhodopsin = visual receptor • Many common receptors belong to this same family • Implications for drug selectivity depending on similarity (evolution) • Membrane bound receptors difficult to crystallise • X-Ray structure of bacteriorhodopsin solved - bacterial protein similar to rhodopsin • Bacteriorhodopsin structure used as ‘template’ for other receptors • Construct model receptors based on template and amino acid sequence • Leads to model binding sites for drug design • Crystal structure for rhodopsin now solved - better template ©1 3. G-protein-coupled receptors (7-TM receptors) 3.4 Bacteriorhodopsin & rhodopsin family Common ance stor Monoamines muscarinic beta alpha Opsins, Rhodopsins Bradyk inin, Endothelins Angiote nsin.Tachykinins Inte rleuk in-8 2 4 5 Mu scarin ic Rec eptor types 3 1 H1 H2 Histamine 1 2A 2B 2C D4 D3 D2 D1A D1B D5 -Adre ne rgic Dopamine rgic 3 2 Rec eptor sub-types 1 -Adre ne rgic ©1 3. G-protein-coupled receptors (7-TM receptors) 3.5 Receptor types and subtypes Reflects differences in receptors which recognise the same ligand Receptor Types Subtypes Adrenergic Alpha () Beta () 1, 2A, 2B, 2C 1, 2, 3 Cholinergic Nicotinic Muscarinic M1-M5 ©1 3. G-protein-coupled receptors (7-TM receptors) 3.5 Receptor types and subtypes • Receptor types and subtypes not equally distributed amongst tissues. • Target selectivity leads to tissue selectivity Heart muscle - 1 adrenergic receptors Fat cells - 3 adrenergic receptors Bronchial muscle - 1& 2 adrenergic receptors GI-tract - 1 2 & 2 adrenergic receptors ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway a) Interaction of receptor with Gs-protein GS-Protein - membrane bound protein of 3 subunits (, , g) - S subunit has binding site for GDP -GDP bound non covalently g GDP ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway a) Interaction of receptor with Gs-protein Ligand Cell membrane Receptor ß g Ligand binding Induced fit G-protein binds ß g Induced fit for G-protein G Protein GDP Binding site for G-protein opens = GDP ß g GTP G-Protein alters shape GDP binding site distorted GDP binding weakened GDP departs ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway a) Interaction of receptor with Gs-protein ß g GTP binds Binding site recognises GTP g ß g Fragmentation and release ß Induced fit G-protein alters shape Complex destabilised • Process repeated for as long as ligand bound to receptor • Signal amplification - several G-proteins activated by one ligand • s Subunit carries message to next stage ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway b) Interaction of s with adenylate cyclase Binding site for s subunit GTP GDP s-subunit Adenylate cyclase GTP hydrolysed to GDP catalysed by s subunit Binding Induced fit Active site (closed) P ATP cyclic AMP Active site (open) Signal transduction (con) s Subunit recombines with ,g dimer to reform Gs protein ATP cyclic AMP Active site (closed) s Subunit changes shape Weaker binding to enzyme Departure of subunit Enzyme reverts to inactive state ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway b) Interaction of s with adenylate cyclase • • • • Several 100 ATP molecules converted before s-GTP deactivated Represents another signal amplification Cyclic AMP becomes next messenger (secondary messenger) Cyclic AMP enters cell cytoplasm with message NH2 NH2 N N O O O HO P O P O P OH OH N N O Adenylate cyclase H O H H O H OH N N O OH ATP N N P OH O H H Cyclic AMP O OH OH ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway c) Interaction of cyclic AMP with protein kinase A (PKA) • Protein kinase A = serine-threonine kinase • Activated by cyclic AMP • Catalyses phosphorylation of serine and threonine residues on protein substrates • Phosphate unit provided by ATP O O H N C Protein kinase A H N H N C H H OH O C HC H OH CH3 Serine P HO O O O Threonine Protein kinase A H N C HC H O CH3 HO P O OH OH Link ©1 Kinase: Any of various enzymes that catalyze the transfer of a phosphate group from a donor, such as ADP or ATP, to an acceptor. Phosphatase: Any of numerous enzymes that catalyze the hydrolysis of esters of phosphoric acid and are important in the absorption and metabolism of carbohydrates, nucleotides, and phospholipids and in the calcification of bone. Phosphorylase: An enzyme that catalyzes the production of glucose phosphate from glycogen and inorganic phosphate. ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway c) Interaction of cyclic AMP with protein kinase A (PKA) Adenylate cyclase ATP cyclic AMP Activation Protein kinase P Link Enzyme (inactive) Enzyme (active) Chemical reaction ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway c) Interaction of cyclic AMP with protein kinase A (PKA) Protein kinase A - 4 protein subunits - 2 regulatory subunits (R) and 2 catalytic subunits (C) cAMP C catalytic subunit C R cAMP binding sites Link R R C R C catalytic subunit Note Cyclic AMP binds to PKA Induced fit destabilises complex Catalytic units released and activated ©1 3. G-protein-coupled receptors (7-TM receptors) 3.6 Signal transduction pathway c) Interaction of cyclic AMP with protein kinase A (PKA) C P Protein + ATP Protein + ADP Phosphorylation of other proteins and enzymes Signal continued by phosphorylated proteins Further signal amplification ©1 3. G-protein-coupled receptors (7-TM receptors) 3.7 Glycogen metabolism - triggered by adrenaline in liver cells Adrenaline s s -Adrenoreceptor adenylate cyclase cAMP Glycogen synthase (active) Protein kinase A Inhibitor (inactive) Catalytic C subunit of PKA Glycogen synthase-P (inactive) Phosphorylase kinase (inactive) Inhibitor-P (active) Phosphatase (inhibited) Phosphorylase kinase-P (active) Phosphorylase b (inactive) Phosphorylase a (active) Glycogen Glucose-1-phosphate ©1 3. G-protein-coupled receptors (7-TM receptors) 3.7 Glycogen metabolism - triggered by adrenaline in liver cells Coordinated effect - activation of glycogen metabolism - inhibition of glycogen synthesis Adrenaline has different effects on different cells - activates fat metabolism in fat cells ©1 3. G-protein-coupled receptors (7-TM receptors) 3.8 GI proteins • Binds to different receptors from those used by Gs protein • Mechanism of activation by splitting is identical • I subunit binds adenylate cyclase to inhibit it • Adenylate cyclase under dual control (brake/accelerator) • Background activity due to constant levels of s and i • Overall effect depends on dominant G-Protein • Dominant G-protein depends on receptors activated ©1 3. G-protein-coupled receptors (7-TM receptors) 3.9 Phosphorylation • Prevalent in activation and deactivation of enzymes • Phosphorylation radically alters intramolecular binding • Results in altered conformations NH3 NH3 NH3 O O O H O Active site closed P O O O O O P O O O Active site open ©1 3. G-protein-coupled receptors (7-TM receptors) 3.10 Drugs interacting with cyclic AMP signal transduction Cholera toxin - constant activation of c.AMP - diahorrea Theophylline and caffeine - inhibit phosphodiesterases - phosphodiesterases responsible for metabolising cyclic AMP - cyclic AMP activity prolonged O O H3C H N H3C N CH3 N N N N O N CH3 Theophylline O N CH3 Caffeine ©1 3. G-protein-coupled receptors (7-TM receptors) 3.11 Signal transduction involving phospholipase C (PLC) • • • • • Gq proteins - interact with different receptors from GS and GI Split by same mechanism to give q subunit q Subunit activates or deactivates PLC (membrane bound enzyme) Reaction catalysed for as long as q bound - signal amplification Brake and accelerator Active site (open) Active site (closed) PLC DG PLC PLC PIP2 IP3 GTP hydrolysis Phosphate DG PLC PIP2 IP3 Binding weakened Active site (closed) q departs PLC enzyme deactivated ©1 3. G-protein-coupled receptors (7-TM receptors) 3.11 Signal transduction involving phospholipase C (PLC) R O C R C O O O CH2 CH O P H CH2 PLC O O P O P HO O H O H HO OH H H + H O P R R O C O C O O CH2 CH OH O P HO HO OH IP3 CH2 DG O P PIP2 Phosphatidylinositol diphosphate (integral part of cell membrane) R= long chain hydrocarbons Inositol triphosphate (polar and moves into cell cytoplasm) P = PO3 2- Diacylglycerol (remains in membrane) ©1 3. G-protein-coupled receptors (7-TM receptors) 3.12 Action of diacylglycerol • • • • • Activates protein kinase C (PKC) PKC moves from cytoplasm to membrane Phosphorylates enzymes at Ser & Thr residues Activates enzymes to catalyse intracellular reactions Linked to inflammation, tumour propagation, smooth muscle activity etc Cell membrane DG Binding site for DG DG DG PKC Active site closed PKC Cytoplasm PKC moves to membrane Cytoplasm DG binds to DG binding site PKC Enzyme (inactive) Cytoplasm Enzyme (active) Chemical reaction Induced fit opens active site ©1 3. G-protein-coupled receptors (7-TM receptors) 3.12 Action of diacylglycerol Drugs inhibiting PKC - potential anti cancer agents CHCO2Me CH3CH2CH2 CH CH CH Me CH C Me O O H O MeO2C CH Me H OH C Me O O H OH O O HO O H Me H Me C O OH H Bryostatin (from sea moss) ©1 3. G-protein-coupled receptors (7-TM receptors) 3.13 Action of inositol triphosphate • IP3 - hydrophilic and enters cell cytoplasm • Mobilises Ca2+ release in cells by opening Ca2+ ion channels • Ca2+ activates protein kinases • Protein kinases activate intracellular enzymes • Cell chemistry altered leading to biological effect ©1 3. G-protein-coupled receptors (7-TM receptors) 3.13 Action of inositol triphosphate Cell membrane IP3 Cytoplasm Calmodulin Calcium stores Calmodulin Ca++ Activation Protein kinase Enzyme (inactive) Ca++ Activation P Enzyme (active) Chemical reaction Protein kinase Enzyme (inactive) P Enzyme (active) Chemical reaction ©1 3. G-protein-coupled receptors (7-TM receptors) 3.14 Resynthesis of PIP2 IP3 + DG several steps PIP2 Inhibition Li+ salts Lithium salts used vs manic depression ©1