run
advertisement

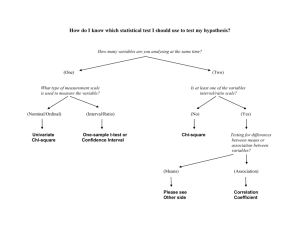
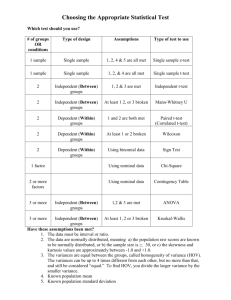
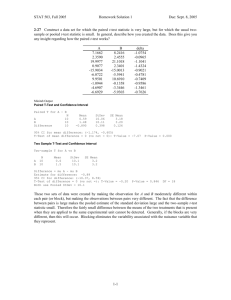
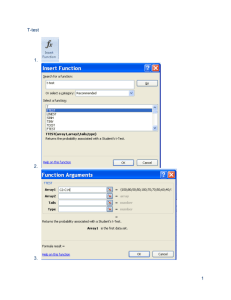
Lecture 9 Raul Cruz-Cano EPIB 698E Fall 2013 Change of Schedule • • • • • • 11/13/2013: Lecture 9-Hypothesis testing 11/20/2013: Lecture 10-Regression 11/27/2013: Review of Midterm 12/4/2013: Lecture 11-Collinearity & Normality Tests 12/11/2013: Lecture 12-Macros 12/18/2013: Final Exam No review before the Final Exam One-Sample T-test 1. A one-sample t-test is used to compare a sample to an average or general population. 2. You may know the average height of men in the U.S., and you could test whether a sample of professional basketball players differ significantly in height from the general U.S. population. 3. A significant difference would indicate that basketball players belong to a different distribution of heights than the general U.S. population. Student's t-test • Independent One-Sample t-test • This equation is used to compare one sample mean to a specific value μ0. t X 0 s/ N • Where s is the grand standard deviation of the sample. N is the sample size. The degrees of freedom used in this test is N-1. 4 T-Test using PROC Univariate • We can also specify a null hypothesis value for the mean when using Proc Univariate by using the mu0 option. proc univariate data=blood mu0=15; var WBC; run; DATA blood; INFILE ‘C:\blood.txt'; INPUT ID Sex $ BloodType $ AgeGroup $ RBC WBC cholesterol; run; Or we can use the SAS Dataset PROC TTEST The following statements are available in PROC TTEST. PROC TTEST < options > ; CLASS variable ; PAIRED variables ; BY variables ; VAR variables ; RUN; PROC TTEST OPTIONS : ALPHA=p specifies that confidence intervals are to be 100(1-p)% confidence intervals, where 0<p<1. By default, PROC TTEST uses ALPHA=0.05. If p is 0 or less, or 1 or more, an error message is printed. H0=m requests tests against m instead of 0 in all three situations (one-sample, twosample, and paired observation t tests). By default, PROC TTEST uses H0=0. DATA=SAS-data-set names the SAS data set for the procedure to use *One sample ttest*; Proc ttest data =blood H0=200; var cholesterol; run; One sample t test Output The TTEST Procedure Variable: cholesterol N Mean Std Dev 795 201.4 49.8867 Mean 201.4 95% CL Mean 198.0 204.9 Std Err 1.7693 Minimum 17.0000 Std Dev 49.8867 Maximum 331.0 95% CL Std Dev 47.5493 52.4676 DF t Value Pr > |t| 95%CL Mean is 95% confidence interval for the mean. 794 0.81 0.4175 95%CL Std Dev is 95% confidence interval for the standard deviation. One sample t test Output N 795 It is the Maximum probability of 331.0 observing a greater absolute 95% CL Mean Std Dev 95% CL Std Dev value of t under the null 198.0 204.9 49.8867 47.5493 52.4676 hypothesis. Mean 201.4 Mean 201.4 Variable: cholesterol Std Dev Std Err Minimum 49.8867 1.7693 17.0000 DF t Value Pr > |t| 794 0.81 0.4175 DF - The degrees of freedom for the t-test is simply the number of valid observations minus 1. We loose one degree of freedom because we have estimated the mean from the sample. We have used some of the information from the data to estimate the mean; therefore, it is not available to use for the test and the degrees of freedom accounts for this T value is the tstatistic. It is the ratio of the difference between the sample mean and the given number to the standard error of the mean. Matched Pairs T-test 1. A matched pairs t-test usually involves the same subjects being measured on some factor at two points in time. 2. For example, subjects could be tested on short-term memory, receive a brief tutorial on memory aids, then have their short-term memory re-tested. 3. A significant difference in score (after-before) would indicate that the tutorial had an effect. Student's t-test • Dependent t-test is used when the samples are dependent; that is, when there is only one sample that has been tested twice (repeated measures) or when there are two samples that have been matched or "paired". t X D 0 sD / N • For this equation, the differences between all pairs must be calculated. The pairs are either one person's pretest and posttest scores or one person in a group matched to another person in another group. The average (XD) and standard deviation (sD) of those differences are used in the equation. The constant μ0 is non-zero if you want to test whether the average of the difference is significantly different than μ0. The degree of freedom used is N-1. 12 Paired Statements • PAIRED: the PAIRED statement identifies the variables to be compared in paired t test 1. You can use one or more variables in the PairLists. 2. Variables or lists of variables are separated by an asterisk (*) or a colon (:). 3. The asterisk (*) requests comparisons between each variable on the left with each variable on the right. 4. Use the PAIRED statement only for paired comparisons. 5. The CLASS and VAR statements cannot be used with the PAIRED statement. title 'Paired Comparison'; data pressure; input SBPbefore SBPafter @@; diff_BP=SBPafter-SBPbefore ; datalines; 120 128 124 131 130 131 118 127 140 132 128 125 140 141 135 137 126 118 130 132 126 129 127 135 ; run; proc ttest data=pressure; paired SBPbefore*SBPafter; run; Paired t test Output The TTEST Procedure Mean of the differences Difference: SBPbefore - SBPafter N Mean Std Dev 12 -1.8333 5.8284 Mean -1.8333 Std Err 1.6825 Minimum -9.0000 Maximum 8.0000 95% CL Mean Std Dev 95% CL Std Dev -5.5365 1.8698 5.8284 4.1288 9.8958 DF t Value Pr > |t| T statistics for testing if the mean of the difference is 0 11 -1.09 0.2992 P =0.3, suggest the mean of the difference is equal to 0 Paired T-test (Example 2) 10 dieters following Atkin’s diet vs. 10 dieters following Jenny Craig Hypothetical RESULTS: Atkin’s group loses an average of 34.5 lbs. J. Craig group loses an average of 18.5 lbs. Conclusion: Atkin’s is better? What if data were paired? e.g., one-to-one matching; find pairs of study participants who have same age, gender, socioeconomic status, degree of overweight, etc. Atkin’s • +4, +3, 0, -3, -4, -5, -11, -14, -15, -300 J. Craig • -8, -10, -12, -16, -18, -20, -21, -24, -26, -30 Enter data differently in SAS… 10 pairs, rather than 20 individual observations data paired; input lossa lossj; diff=lossa-lossj; datalines ; +4 -8 +3 -10 0 -12 -3 -16 -4 -18 -5 -20 -11 -21 -14 -24 -15 -26 -300 -30 ; run; Tests in SAS… /*to get all paired tests*/ proc univariate data=paired; var diff; run; /*To get just paired ttest*/ proc ttest data=paired; var diff; run; /*To get paired ttest, alternatively*/ proc ttest data=paired; paired lossa*lossj; run; Two-Sample T-test 1. A two-sample t-test compares two groups on some factor. 2. For example, one group could receive an experimental treatment and the second group could receive a standard of care treatment or placebo. 3. Notice that in a two-sample t-test, two distinct groups are being compared, as opposed to the onesample, where one group is compared to a general average, or a matched-pairs, where only one group is being measured twice. Two independent samples t-test • An independent samples t-test is used when you want to compare the means of a normally distributed interval dependent variable for two independent groups. For example, using the hsb2 data file, say we wish to test whether the mean for write is the same for males and females. proc ttest data = "c:\hsb2"; class female; var write; run; CLASS: CLASS statement giving the name of the classification (or grouping) variable must accompany the PROC TTEST statement in the two independent sample cases (TWO SAMPLE T TEST). The class variable must have two, and only two, levels. Two Independent Samples: Distribution Free Tests • There are times when the assumptions for using a ttest are not met. • One common problem is that the data are not normally distributed, and your sample size is small. • Another common problem is that the data values may only represent ordered categories. • We need a nonparametric test to analyze differences in central tendencies for ordinal data. • For very small samples, nonparametric tests are often more appropriate since assumptions concerning distributions are difficult to determine. Distribution Free Tests • The biggest difference between a parametric and nonparametric test is the fact that a parametric test assumes that the data under investigation is coming from a normal distribution. • The SAS software provides several nonparametric tests such as the Wilcoxon rank-sum test and the Kruskal-Wallis test when dealing with two or more samples. Non-parametric tests • t-tests require your outcome variable to be normally distributed (or close enough). • Non-parametric tests are based on RANKS instead of means and standard deviations (=“population parameters”). Example: non-parametric tests 10 dieters following Atkin’s diet vs. 10 dieters following Jenny Craig Hypothetical RESULTS: Atkin’s group loses an average of 34.5 lbs. J. Craig group loses an average of 18.5 lbs. Conclusion: Atkin’s is better? Enter data in SAS… data nonparametric; input loss diet $; datalines ; +4 atkins +3 atkins 0 atkins -3 atkins -4 atkins -5 atkins -11 atkins -14 atkins -15 atkins -300 atkins -8 jenny -10 jenny -12 jenny -16 jenny -18 jenny -20 jenny -21 jenny -24 jenny -26 jenny -30 jenny ; run; t-test doesn’t work… • Comparing the mean weight loss of the two groups is not appropriate here. • The distributions do not appear to be normally distributed. • Moreover, there is an extreme outlier (this outlier influences the mean a great deal). Statistical tests to compare ranks: • Wilcoxon rank-sum test is analogue of twosample t-test. • Wilcoxon signed-rank test is analogue of onesample t-test, usually used for paired data NPAR1WAY Procedure • The NPAR1WAY procedure provides the following location tests: Wilcoxon rank sum test (Mann-Whitney U test), Median test, Savage test, and Van der Waerden test. • Also note that the Wilcoxon rank sum test can be obtained from the FREQ procedure. Wilcoxon rank-sum test • RANK the values, 1 being the least weight loss and 20 being the most weight loss. • Atkin’s • +4, +3, 0, -3, -4, -5, -11, -14, -15, -300 • 1, 2, 3, 4, 5, 6, 9, 11, 12, 20 • J. Craig • -8, -10, -12, -16, -18, -20, -21, -24, -26, -30 • 7, 8, 10, 13, 14, 15, 16, 17, 18, 19 Wilcoxon “rank-sum” test • Sum of Atkin’s ranks: • 1+ 2 + 3 + 4 + 5 + 6 + 9 + 11+ 12 + 20=73 • Sum of Jenny Craig’s ranks: 7 + 8 +10+ 13+ 14+ 15+16+ 17+ 18+19=137 • Jenny Craig clearly ranked higher! Wilcoxon rank-sum (Example 1) /*to get wilcoxon rank-sum test*/ proc npar1way wilcoxon data=nonparametric; class diet; var loss; run; Compare p-values /*To get ttest*/ 0.0156 vs. 0.5962 proc ttest data=nonparametric; class diet; var loss; run; Wilcoxon rank-sum test for two samples (Example 2) 1. 2. Consider the following experiment. We have two groups, A and B. Group B has been treated with a drug to prevent tumor formation. Both groups are exposed to a chemical that encourages tumor growth. The masses (in grams) of tumors in groups A and B are: DATA TUMOR; INPUT GROUP $ MASS @@; DATALINES ; A 3.1 A 2.2 A 1.7 A 2.7 A 2.5 B 0.0 B 0.0 B 1.0 B 2.3 ; PROC NPAR1WAY DATA =TUMOR WILCOXON; TITLE 'NONPARAMETRIC TEST TO COMPARE TUMOR MASSES’ ; CLASS GROUP; VAR MASS; RUN; Wilcoxon rank-sum test for two samples (Example 3) • Consider the following example, Researcher B is interested in testing the difference between the effectiveness of two allergy drugs out on the market. • He would like to administer drug A to a random sample of study subjects and then drug B to another random sample who suffer from the same symptoms as those individuals taking drug A. • Researcher B would like to see if there is a difference between the two groups in the time, in minutes, for subjects to feel relief from their allergy symptoms. Wilcoxon rank-sum test for two samples (Example 3) data drugtest; input subject drug_group $ time; datalines; 1 A 43 2 A 40 3 A 32 4 A 37 5 A 55 6 A 50 7 A 52 8 A 33 9 B 28 10 B 33 11 B 48 12 B 37 13 B 40 14 B 42 15 B 35 16 B 43 ; run; proc means median min max; by drug_group; var time; run; proc npar1way wilcoxon; class drug_group; var time; run; The p-values (.3431) are above 0.05, you cannot reject the null hypothesis and must conclude that there is no difference between the median times to relief for both drug groups. Wilcoxon “signed-rank” test H0: median weight loss in Atkin’s group = 0 Ha:median weight loss in Atkin’s not 0 Atkin’s • +4, +3, 0, -3, -4, -5, -11, -14, -15, -300 Rank absolute values of differences (ignore zeroes): Ordered values: 300, 15, 14, 11, 5, 4, 4, 3, 3, 0 Ranks: 1 2 3 4 5 6-7 8-9 Sum of negative ranks: 1+2+3+4+5+6.5+8.5=30 Sum of positive ranks: 6.5+8.5=15 Signed-rank (Example 1) /*to get one-sample tests (both student’s t and signed-rank*/ proc univariate data=nonparametric; var loss; where diet="atkins"; run; Compare p-values You need to use the option ‘m0=’ to change the alternative hypothesis, not ‘h0=’ as in PROC TTEST ANOVA • A one-way analysis of variance (ANOVA) is used when you have a categorical independent variable (with two or more categories) and a normally distributed interval dependent variable and you wish to test for differences in the means of the dependent variable broken down by the levels of the independent variable. • Just an extension of the t-test (an ANOVA with only two groups is mathematically equivalent to a t-test). ANOVA (ANalysis Of VAriance) • Idea: For two or more groups, test difference between means, for quantitative normally distributed variables. • Like the t-test, ANOVA is “parametric” test—assumes that the outcome variable is roughly normally distributed The “F-test” Is the difference in the means of the groups more than background noise (=variability within groups)? Variabilit y between groups F Variabilit y within groups Spine bone density vs. menstrual regularity 1.2 1.1 1.0 S P I N E 0.9 Within group variability Between group variation Within group variability Within group variability 0.8 0.7 amenorrheic oligomenorrheic eumenorrheic The F-distribution • A ratio of sample variances follows an Fdistribution: 2 between 2 within The F ~ Fn , m F-test tests the hypothesis that two sample variances are equal. will be close to 1 if sample variances are equal. 2 2 H 0 : between within H a : 2 between 2 within The F-distribution • The F-distribution is a continuous probability distribution that depends on two parameters n and m (numerator and denominator degrees of freedom, respectively): ANOVA Table Source of variation Between (k groups) d.f. Sum of squares k-1 SSB Mean Sum of Squares SSB/k-1 (sum of squared deviations of group means from F-statistic SSB SSW p-value Go to k 1 nk k Fk-1,nk-k chart grand mean) Within nk-k (n individuals per group) Total variation nk-1 SSW (sum of squared deviations of observations from their group mean) s2=SSW/nk-k TSS (sum of squared deviations of observations from grand mean) TSS=SSB + SSW ANOVA summary • A statistically significant ANOVA (F-test) only tells you that at least two of the groups differ, but not which ones differ. • Determining which groups differ (when it’s unclear) requires more sophisticated analyses to correct for the problem of multiple comparisons… ANOVA • The following example studies the effect of bacteria on the nitrogen content of red clover plants. The treatment factor is bacteria strain, and it has six levels. Five of the six levels consist of five different Rhizobium trifolii bacteria cultures combined with a composite of five Rhizobium meliloti strains. The sixth level is a composite of the five Rhizobium trifolii strains with the composite of the Rhizobium meliloti. Red clover plants are inoculated with the treatments, and nitrogen content is later measured in milligrams. title1 'Nitrogen Content of Red Clover Plants'; data Clover; input Strain $ Nitrogen @@; datalines; 3DOK1 19.4 3DOK1 32.6 3DOK1 27.0 3DOK1 32.1 3DOK1 33.0 3DOK5 17.7 3DOK5 24.8 3DOK5 27.9 3DOK5 25.2 3DOK5 24.3 3DOK4 17.0 3DOK4 19.4 3DOK4 9.1 3DOK4 11.9 3DOK4 15.8 3DOK7 20.7 3DOK7 21.0 3DOK7 20.5 3DOK7 18.8 3DOK7 18.6 3DOK13 14.3 3DOK13 14.4 3DOK13 11.8 3DOK13 11.6 3DOK13 14.2 COMPOS 17.3 COMPOS 19.4 COMPOS 19.1 COMPOS 16.9 COMPOS 20.8 ; run; proc anova data = Clover; class strain; model Nitrogen = Strain; run; proc freq data = Clover; tables Strain; run; ANOVA Graphs ods graphics on; proc anova data = Clover; class strain; model Nitrogen = Strain; run; ods graphics off; ANOVA • The test for Strain suggests that there are differences among the bacterial strains, but it does not reveal any information about the nature of the differences. Mean comparison methods can be used to gather further information. Another ANOVA Example • Let’s assume that the researcher has data on individuals from three different diet camps. • All the researcher is concerned with is seeing whether the mean weights of the individuals in each camp are significantly different from one another. • Since we are comparing three different means, we must employ the use of ANOVA. Another Example data expanova; input group weight; datalines; 1 223 1 234 1 254 1 267 1 234 2 287 2 213 2 215 2 234 2 256 3 234 3 342 3 198 3 256 3 303 ; proc anova; class group; model weight = group; means group; run; Adds little summary at the end Non-parametric ANOVA Kruskal-Wallis one-way ANOVA Extension of the Wilcoxon Sign-Rank test for 2 groups; based on ranks Proc NPAR1WAY in SAS Kruskal-Wallis (Example) 1. The data consist of weight gain measurements for five different levels of gossypol additive. 2. Gossypol is a substance contained in cottonseed shells, and these data were collected to study the effect of gossypol on animal nutrition. data Gossypol; input Dose n; do i=1 to n; input Gain @@; output; end; datalines; 0 16 228 229 218 216 224 208 235 229 233 219 224 220 232 200 208 232 .04 11 186 229 220 208 228 198 222 273 216 198 213 .07 12 179 193 183 180 143 204 114 188 178 134 208 196 .10 17 130 87 135 116 118 165 151 59 126 64 78 94 150 160 122 110 178 .13 11 154 130 130 118 118 104 112 134 98 100 104 ; run; Kruskal-Wallis (Example) proc npar1way data=Gossypol; class Dose; var Gain; run; 1. The p-value, or probability of a larger statistic under the null hypothesis, is <.0001. 2. This leads to rejection of the null hypothesis that there is no difference in location for Gain among the levels of Dose