Second presentation - DIMACS REU

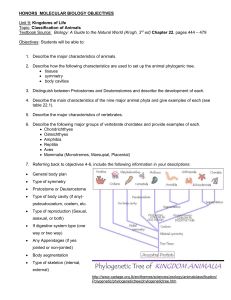
DNA PACKING:
Distances Between
DNA Molecules in Crystals
Bryson W. Finklea
St. John's College
DIMACS REU
Unit Cell
Asymmetric Unit Unit Cell 3x3x3 Block of Unit Cells
Asymmetric Unit Unit Cell 3x3x3 Block of Unit Cells
(Rasmol images)
Space Group: C 2 NDB #: bdl042
Asymmetric Unit Unit Cell 3x3x3 Block of Unit Cells
(Rasmol images)
Space Group: C 2 NDB #: bdl042
Asymmetric Unit Unit Cell 3x3x3 Block of Unit Cells
(Rasmol images)
Space Group: P 6
1
NDB #: adh007
Asymmetric Unit Unit Cell 3x3x3 Block of Unit Cells
(Rasmol images)
Space Group: P 6
1
NDB #: adh007
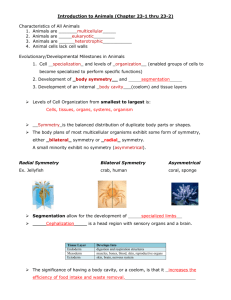
Example of 2D symmetry in a wallpaper pattern
To show symmetry:
● pick a point
● find all equivalent points
Example of 2D symmetry in a wallpaper pattern
●
Connecting 4 lattice points in a parallelogram gives a unit cell
●
Unit cell – the basic unit that repeats in every direction
●
Some unit cells are conventionally used because of their higher symmetry
Example of 2D symmetry in a wallpaper pattern
●
Unit cell*
●
Asymmetric Unit –the simplest unit on which the symmetry operations can act to produce the entire symmetrical structure*
Reflection Axis
Glide Reflection Axis
90° Rotation Point
180° Rotation Point
Symmetry elements of this wallpaper group
* Although the spirit of what I show is correct, it appears from the following website that my choice of conventional unit cell and choice of asymmetric unit may be unconventional or even wrong. See the last example in the n=4 section of the following website: http://jwilson.coe.uga.edu/EMT668/EMAT6680.F99/McCallum/WALLPA~1/SEVENT~1.HTM
3D Symmetry
(Unknown)
Generalized 3D unit cell —a parallelepiped
Categories of Space Groups
http://www.chem.ox.ac.uk/icl/heyes/structure_of_solids/Lecture1/Bravais.gif
Proper symmetry elements in 3D:
•Rotation axes (by 60°, 90°, 120°, or 180°)
Notated: 6, 4, 3, and 2, respectively
•Screw Axes (translation and rotation)
Notated: 6
1
•(Translation)
, 6
2
, 6
3
, 6
4
, 6
5
; 4
1
, 4
2
, 4
3
; 3
1
, 3
2
; and 2
1
Proper symmetry elements in 3D:
•Reflection planes
•Glide reflection planes (reflection and translation)
•Inversion points
•Rotary inversion axes (rotation and inversion)
Symmetry operations –the actual changes carried out in relation to a symmetry element
Sets of symmetry operations form algebraic groups called space groups .
•230 space groups
Space Group Total Total w/ DNA DNA DNA
Entries Res<2.0
only complexes Total
C 1 2 1
P 21 21 21
135
476
67
222
17
126
75 92
324 450
H 3
52 Others
24
984
10
317 164
5 15 20
534 698
54 (of 230) Total 1619 616 312 948 1260
Asymmetric Unit Unit Cell 3x3x3 Block of Unit Cells
(Rasmol images)
Space Group: C 2 NDB #: bdl042
My Project
Characterizing Intermolecular Contacts of DNA
Data from Nucleic Acid Database (NDB):
● orthogonal coordinates of atoms in asymmetric unit
● conversion matrix from orthogonal to fractional (unit cell) coordinates
● equivalent positions in equation form (info from symmetry elements)
● unit cell dimensions and angles
To revise a computer program to:
● reconstruct coordinates of the atoms in a unit cell
●
…then in a 3x3x3 block of unit cells
●
Measure distances between pairs of atoms in different molecules
Example output from program
(for bdl042.pdb)
UCELL MOL AT# AT RES UCELL MOL AT# AT RES DISTANCE
0. 0. 0. 1 216 O2 C 0. 0. 0. 2 227 C4* G 2.93
0. 0. 0. 1 240 C2 G 0. 0. 0. 2 288 O4* T 2.95
0. 0. 0. 1 264 C5* G 0. 0. 0. 2 264 C5* G 2.96
0. 0. 0. 1 288 O4* T 0. 0. 0. 2 240 C2 G 2.95
0. 0. 0. 1 19 O2P G 1. 0. 1. 2 19 O2P G 2.75
0. 0. 0. 1 37 N3 G 1. 0. 1. 2 484 N2 G 2.98
0. 0. 0. 1 452 O4* C 1. 0. 1. 2 473 O3* G 2.62
0. 0. 0. 1 454 O3* C 1. 0. 1. 2 469 C5* G 2.67
0. 0. 0. 1 469 C5* G 1. 0. 1. 2 454 O3* C 2.67
0. 0. 0. 1 469 C5* G 1. 0. 1. 2 469 C5* G 2.28
0. 0. 0. 1 471 O4* G 1. 0. 1. 2 471 O4* G 2.55
0. 0. 0. 1 473 O3* G 1. 0. 1. 2 452 O4* C 2.62
0. 0. 0. 1 484 N2 G 1. 0. 1. 2 37 N3 G 2.98
0. 0. 0. 1 484 N2 G 1. 0. 1. 2 484 N2 G 2.73
0. 0. 0. 2 19 O2P G -1. 0.-1. 1 19 O2P G 2.75
0. 0. 0. 2 37 N3 G -1. 0.-1. 1 484 N2 G 2.98
0. 0. 0. 2 452 O4* C -1. 0.-1. 1 473 O3* G 2.62
0. 0. 0. 2 454 O3* C -1. 0.-1. 1 469 C5* G 2.67
0. 0. 0. 2 469 C5* G -1. 0.-1. 1 454 O3* C 2.67
0. 0. 0. 2 469 C5* G -1. 0.-1. 1 469 C5* G 2.28
0. 0. 0. 2 471 O4* G -1. 0.-1. 1 471 O4* G 2.55
0. 0. 0. 2 473 O3* G -1. 0.-1. 1 452 O4* C 2.62
0. 0. 0. 2 484 N2 G -1. 0.-1. 1 37 N3 G 2.98
0. 0. 0. 2 484 N2 G -1. 0.-1. 1 484 N2 G 2.73
Background
Nature
(Human Genome Project Information of the DOE)
Nanotechnology
(Dr. Nadrian Seeman, Department of Chemistry,
New York University)
Future Work:
●
Tweak output features of program
●
Run program on all DNA-only files of NDB
●
Characterize intermolecular contacts in different ways e.g. angle between DNA molecular axes
●
Alter notation of program to run it on NDB files containing RNA, protein, and drug molecules
Acknowledgments
DIMACS REU
NSF Support
Advisor:
Wilma Olson, Department of Chemistry,
Rutgers University
Additional Advisor:
A.R. Srinivasan, Department of Chemistry,
Rutgers University
(background: http://www.karolinskaeducation.ki.se/services/courses/selection_courses_se.html)