Mumps' Presentation
advertisement

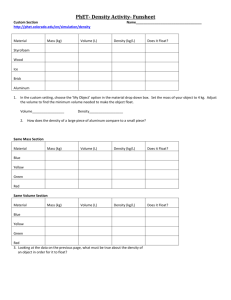
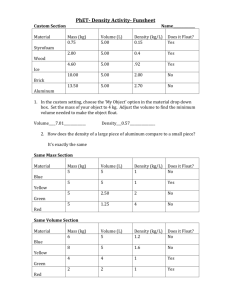
Calculating Measures of Comorbidity Using Administrative Data Vicki Stagg Statistical Programmer Department of Community Health Sciences Dr. Robert Hilsden Associate Professor Departments of Medicine and Community Health Sciences Dr. Hude Quan Associate Professor Centre for Health and Policy Studies (CHAPS) Department of Community Health Sciences University of Calgary Calgary, Alberta, Canada Background • Medical Administrative Data • Inpatient hospital visit information • Comorbidity • Pre-existing diagnosis / additional complication of admitted patient • Comorbidity Index • For measurement of burden of disease and case-mix adjustment • Allows for stratification or adjustment by severity of illness • Two common tools – Charlson and Elixhauser • Clinical Conditions • International Classification of Disease – 9th Revision, Clinical Modification (ICD-9-CM codes) – 10th Revision (ICD-10 codes) (International statistical classification of disease and related health problems, 1992) Algorithms included in ado programs • Charlson (17 comorbidity definitions) • Presence/absence, weighted sum (Charlson index) • Charlson index developed to predict risk of one-year mortality from comorbid illness (J Chron Dis, 1987;40(5):373-383) • Deyo modification for ICD-9-CM (J Clin Epi, 1992;45(6):613-619) • Quan’s Enhanced ICD-9-CM • Quan’s ICD-10 (Medical Care, 2005;43(11):1130-1139) • Elixhauser (30 comorbidity definitions) • Presence/absence, sum (Medical Care, 1998:36(1):8-27) • Quan’s Enhanced ICD-9-CM • Quan’s ICD-10 (Medical Care, 2005;43(11):1130-1139) Algorithms – development of ICD-10 & enhanced ICD-9-CM • ICD-10 comorbidity coding algorithm • • • Based on Charlson index Swiss, Australian, Canadian collaborative groups ICD-10 Canadian version (ICD-10-CA) • Enhanced ICD-9-CM coding algorithm • • Back-translated from new ICD-10 coding algorithm To improve original Deyo (Charlson) and Elixhauser comorbidity classifications Charlson Comorbidities with Corresponding ICD-9-CM and ICD-10 Codes (Medical Care. 2005;43:1130-1139) Example coding algorithm Comorbidity Deyo Enhanced ICD-10 Myocardial infarction 410.x, 412.x 410.x, 412.x I21.x, I22.x, I25.2 Congestive heart failure 428.x 398.91, 402.01, 402.11, 402.91, 404.01, 404.03, 404.11, 404.13, 404.91, 404.93, 425.4-425.9, 428.x I09.9, I11.0, I13.0, I13.2, I12.5, I42.0, I42.5-I42.9, I43.x, I40.x, P29.0 (Medical Care, 2005;43(11):1130-1139) Charlson Comorbidities & Weights CHARLSON COMORBIDITY ASSIGNED WEIGHTS 1. Myocardial infarction 1 2. Congestive heart failure 1 3. Peripheral vascular disease 1 4. Cerebrovascular disease 1 5. Dementia 1 6. Chronic pulmonary disease 1 7. Rheumatic disease 1 8. Peptic ulcer disease 1 9. Mild liver disease 1 10. Diabetes without chronic complication 1 11. Diabetes with end organ damage 2 12. Hemiplegia / paraplegia 2 13. Renal disease 2 14. Any malignancy/lymphoma/leukemia 2 15. Moderate or severe liver disease 3 16. Metastatic solid tumor 6 17. AIDS/HIV 6 Input data • Patient demographic data • ID variable (string) required if multiple visits • Comorbidity diagnoses codes (strings) • Charlson – ICD-9-CM / ICD-10 • Elixhauser – ICD-9-CM / ICD-10 • Additional medical information • For subsequent modeling, if desired Syntax • Charlson • charlson varlist [if exp] [in range], index(string) [idvar(varname) diagprfx(string) assign0 wtchrl cmorb noshow] by may be used with charlson • Elixhauser • elixhauser varlist [if exp] [in range], index(string) [idvar(varname) diagprfx(string) smelix cmorb noshow] by may be used with elixhauser Input options • index (string) • ICD-9-CM (charlson) • Enhanced ICD-9-CM (charlson/elixhauser) • ICD-10 (charlson/elixhauser) c e 10 • idvar(varname) • Required when multiple records per patient • diagprfx(string) • Gives common root of the comorbidity variables • Necessary only when varlist not used • assign0 • Only applicable to charlson • Flag to apply hierarchical method Output options • wtchrl (charlson command) • Presents summary of Charlson Index (frequencies of weighted sums) • wtelix (elixhauser command) • Displays frequencies of sum of elixhauser comorbidities • cmorb • Displays frequencies of individual comorbidities • noshow • Controls display of chosen options Sample program #1 – charlson (Enhanced ICD-9-CM Algorithm) • Input data (ICD-9-CM codes) Small Sample Data patientid diag1 diag2 id1 39891 id2 0930 id3 2500 2507 id4 342 2500 id5 3441 342 id6 2500 5722 id7 5722 196 id8 042 id9 V427 4561 176 197 id10 5834 diag3 342 3441 V427 Sample program #1 – charlson • Command – . charlson, index(e) diagprfx(diag) wtchrl cmorb Sample program #1 – charlson – Output (part 1) • (Option noshow omitted) (0 observations deleted) COMORBIDITY INDEX MACRO Providing COMORBIDITY INDEX Summary OPTIONS SELECTED: INPUT DATA: Enhanced ICD-9 OBSERVATIONAL UNIT: Visits ID VARIABLE NAME (Given only if Unit is Patients): PREFIX of COMORBIDITY VARIABLES: diag HIERARCHY METHOD APPLIED: NO SUMMARIZE CHARLSON INDEX and WEIGHTS: YES SUMMARIZE INDIVIDUAL COMORBIDITIES: YES Please wait. Thank you! Program takes a few minutes - there are up to 3 ICD codes per subject. Iteration 1 of 3 - Program is running - Please wait Iteration 2 of 3 - Program is running - Please wait Iteration 3 of 3 - Program is running - Please wait Total Number of Observational Units (Visits OR Patients): 10 Sample program #1 – charlson – Output (part 2) CHARLSON | INDEX | Freq. Percent Cum. ------------+----------------------------------1 | 1 10.00 10.00 2 | 1 10.00 20.00 3 | 2 20.00 40.00 4 | 2 20.00 60.00 5 | 1 10.00 70.00 6 | 1 10.00 80.00 9 | 2 20.00 100.00 ------------+----------------------------------Total | 10 100.00 • (option wtchrl) GROUPED | CHARLSON | INDEX | Freq. Percent Cum. ------------+----------------------------------1 | 1 10.00 10.00 2 | 9 90.00 100.00 ------------+----------------------------------Total | 10 100.00 Variable | Obs Mean Std. Dev. Min Max -------------+-------------------------------------------------------charlindex | 10 4.6 2.716207 1 9 Sample program #1 – charlson – Output (part 3) • (option cmorb) • selected comorbidities displayed Diabetes | Freq. Percent Cum. ------------+----------------------------------Absent | 7 70.00 70.00 Present | 3 30.00 100.00 ------------+----------------------------------Total | 10 100.00 Diabetes + | Complicatio | ns | Freq. Percent Cum. ------------+----------------------------------Absent | 9 90.00 90.00 Present | 1 10.00 100.00 ------------+----------------------------------Total | 10 100.00 Output dataset – describe obs: 10 vars: 41 17 Oct 2007 10:08 size: 1,880 (99.9% of memory free) ------------------------------------------------------------------------------storage display value variable name type format label variable label ------------------------------------------------------------------------------id str23 %23s diag1 str5 %9s diag2 str4 %9s diag3 str4 %9s ynch1 float %9.0g ynlab AMI (Acute Myocardial) ynch2 float %9.0g ynlab CHF (Congestive Heart) ynch3 float %9.0g ynlab PVD (Peripheral Vascular) ynch4 float %9.0g ynlab CEVD (Cerebrovascular ynch5 float %9.0g ynlab Dementia ynch6 float %9.0g ynlab COPD (Chronic Obstructive Pulmonary) ynch7 float %9.0g ynlab Rheumatoid Disease ynch8 float %9.0g ynlab PUD (Peptic Ulcer) ynch9 float %9.0g ynlab Mild LD (Liver) ynch10 float %9.0g ynlab Diabetes ynch11 float %9.0g ynlab Diabetes + Complications ynch12 float %9.0g ynlab HP/PAPL (Hemiplegia or Paraplegia) Output dataset – describe continued ynch13 float %9.0g ynlab RD (Renal) ynch14 float %9.0g ynlab Cancer ynch15 float %9.0g ynlab Moderate/Severe LD (Liver) ynch16 float %9.0g ynlab Metastic Cancer ynch17 float %9.0g ynlab AIDS weightch1 float %9.0g weightch2 float %9.0g weightch3 float %9.0g weightch4 float %9.0g weightch5 float %9.0g weightch6 float %9.0g weightch7 float %9.0g weightch8 float %9.0g weightch9 float %9.0g weightch10 float %9.0g weightch11 float %9.0g weightch12 float %9.0g weightch13 float %9.0g weightch14 float %9.0g weightch15 float %9.0g weightch16 float %9.0g weightch17 float %9.0g charlindex float %9.0g CHARLSON INDEX grpci float %9.0g GROUPED CHARLSON INDEX ------------------------------------------------------------------------------Sorted by: Note: dataset has changed since last saved Output dataset – selected variables . list id ynch10 ynch11 ynch15 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. +------------------------------------+ | id ynch10 ynch11 ynch15 | |------------------------------------| | id1 Absent Absent Absent | | id2 Absent Absent Absent | | id3 Present Present Absent | | id4 Present Absent Absent | | id5 Absent Absent Absent | |------------------------------------| | id6 Present Absent Present | | id7 Absent Absent Present | | id8 Absent Absent Absent | | id9 Absent Absent Present | | id10 Absent Absent Absent | +------------------------------------+ . list id weightch10 weightch11 weightch15, c 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. +------------------------------+ | id we~10 we~11 we~15 | |------------------------------| | id1 0 0 0 | | id2 0 0 0 | | id3 1 2 0 | | id4 1 0 0 | | id5 0 0 0 | |------------------------------| | id6 1 0 3 | | id7 0 0 3 | | id8 0 0 0 | | id9 0 0 3 | | id10 0 0 0 | +------------------------------+ Output dataset – Charlson index & grouped Charlson index . list id 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. charlindex grpci +-------------------------+ | id charli~x grpci | |-------------------------| | id1 5 2 | | id2 1 1 | | id3 3 2 | | id4 3 2 | | id5 2 2 | |-------------------------| | id6 4 2 | | id7 9 2 | | id8 6 2 | | id9 4 2 | | id10 9 2 | +-------------------------+ Program rerun with assign0 option – (changes frequencies) . comorbid, index(e) diagprfx(diag) wtchrl cmorb assign0 CHARLSON | INDEX | Freq. Percent Cum. ------------+----------------------------------1 | 1 10.00 10.00 2 | 2 20.00 30.00 3 | 2 20.00 50.00 4 | 1 10.00 60.00 5 | 1 10.00 70.00 6 | 1 10.00 80.00 7 | 1 10.00 90.00 9 | 1 10.00 100.00 ------------+----------------------------------Total | 10 100.00 GROUPED | CHARLSON | INDEX | Freq. Percent Cum. ------------+----------------------------------1 | 1 10.00 10.00 2 | 9 90.00 100.00 ------------+----------------------------------Total | 10 100.00 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. +-------------------------+ | id charli~x grpci | |-------------------------| | id1 5 2 | | id2 1 1 | | id3 2 2 | | id4 3 2 | | id5 2 2 | |-------------------------| | id6 4 2 | | id7 9 2 | | id8 6 2 | | id9 3 2 | | id10 7 2 | +-------------------------+ Selected comorbidities revisitedDiabetes | Freq. Percent Cum. ------------+----------------------------------Absent | 8 80.00 80.00 Present | 2 20.00 100.00 ------------+----------------------------------Total | 10 100.00 Diabetes + | Complicatio | ns | Freq. Percent Cum. ------------+----------------------------------Absent | 9 90.00 90.00 Present | 1 10.00 100.00 ------------+----------------------------------Total | 10 100.00 Sample program #2 – elixhauser (ICD-10 Algorithm) • Input - real inpatient data obs: 2,987 vars: 43 18 Oct 2007 10:18 size: 1,000,645 (90.5% of memory free) ------------------------------------------------------------------------------storage display value variable name type format label variable label ------------------------------------------------------------------------------dx1 str6 %9s DIAG1 dx2 str6 %9s DIAG2 . . . dx24 str6 %9s DIAG24 dx25 str6 %9s DIAG25 cdr_keyforqsh~e long %12.0g CDR_KEY (for QSHI use) admitdate str20 %20s Admit Date dischargedate str20 %20s Discharge Date acutelosdays int %8.0g ACUTE LOS (days) birthdate str11 %11s Birth Date age int %8.0g AGE pc str6 %9s PC residence str7 %9s RESIDENCE entrycodetoho~l str61 %61s ENTRY CODE to hospital strokediagtyp~a str25 %25s Stroke Diag Type when Stroke not the Main Diag gender long %8.0g gender gender site long %8.0g site site stroketype long %13.0g stroke Stroke type disposition long %60.0g disp discharge disposition cohort float %9.0g cohort cohort ------------------------------------------------------------------------------- Sample program #2 – elixhauser • Command – . elixhauser dx1-dx25, index(10) smelix cmorb Sample program #2 – elixhauser Output ELIX | COMORBIDITY | SUM | Freq. Percent Cum. ------------+----------------------------------0 | 402 13.46 13.46 1 | 715 23.94 37.40 2 | 751 25.14 62.54 3 | 529 17.71 80.25 4 | 303 10.14 90.39 5 | 174 5.83 96.22 6 | 71 2.38 98.59 7 | 30 1.00 99.60 8 | 10 0.33 99.93 9 | 1 0.03 99.97 10 | 1 0.03 100.00 ------------+----------------------------------Total | 2,987 100.00 Variable | Obs Mean Std. Dev. Min Max -------------+-------------------------------------------------------elixsum | 2987 2.216605 1.621281 0 10 ELIXHAUSER COMORBIDITY Congestive Heart Failure Cardiac Arrhythmias PERCENT 8.20 22.23 Valvular Disease 4.49 Pulmonary Circulation Disorders 1.77 Peripheral Vascular Disorders 5.49 Hypertension, Uncomplicated 58.49 Paralysis 25.68 Other Neurological Disorders 23.84 Chronic Pulmonary Disease Diabetes, Uncomplicated 7.20 15.80 Diabetes, Complicated 4.08 Hypothyroidism 3.85 Renal Failure 4.69 Liver Disease 0.77 Peptic Ulcer Disease Excluding Bleeding 0.40 AIDS/HIV 0.13 …continued ELIXHAUSER COMORBIDITY PERCENT Lymphoma 0.57 Metastatic Cancer 1.61 Solid Tumor Without Metastasis 2.98 Rheumatoid Arthritis/Collagen Vascular 1.47 Coagulopathy 2.68 Obesity 3.31 Weight Loss 0.67 Fluid and Electrolyte Disorders 6.46 Blood Loss Anemia 0.50 Deficiency Anemia 1.31 Alcohol Abuse 3.35 Drug Abuse 0.74 Psychoses 0.50 Depression 4.59 Hypertension, Complicated 3.82 Acknowledgements I would like to express sincere gratitude to: • Dr. Robert Hilsden Depts. of Medicine/ Community Health Sciences, U of Calgary For supervising this work and for all his advice and support. • Dr. Hude Quan Centre for Health and Policy Studies Dept. of Community Health Sciences, U of Calgary For providing the SAS code and databases and for his support. • Haifeng Zhu MSc Graduate Student Dept. of Community Health Sciences For her assistance with converting the Elixhauser algorithms to Stata. • Malcolm Stagg Student, Vista Virtual School, Calgary AB My son, for his help with preparing this PowerPoint presentation and all his encouragement. • Andrew Stagg Intern, Google Inc., Mountain View CA My son, for his encouragement. SUGGESTIONS / COMMENTS WELCOME vlstagg@ucalgary.ca Thank you!