Splicing - iPlant Pods
advertisement

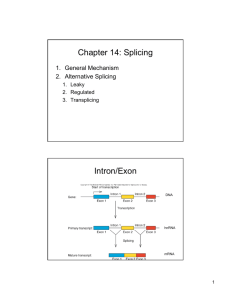
Alternative Splicing mRNA Splicing During RNA processing internal segments are removed from the transcript and the remaining segments spliced together. Internal RNA segments that are removed are named introns; the spliced segments are defined as exons. Splicing prevalent process in eukaryotes (not 100%, though) Language erosion: 1-exon genes Canonical splice sites 5’ Splice Site Intron Exon 3’ Splice Site Pre-mRNA Exon Reddy, S.N. Annu. Rev. Plant Biol. 2007 58:267-94 Of 1588 examined predicted splice sites in Arabidopsis 1470 sites (93%) followed the canonical GT…AG consensus. (Plant (2004) 39, 877–885) An example from A. thaliana Multiple splice variants produced from the same gene Alternative Splicing DNALC Clip: http://dnalc.org/resources/3d/ Removing different segments from mRNAs leads to alternative splice forms of a gene/transcript. Can occur in any part of the transcript including UTRs and can alter start codons, stop codons, reading frame, CDS, UTRs. May alter stability-life, translation (time, location, duration), protein sequence, some or all of the above. Alternative splice forms = Protein isoforms Contributes to protein diversity Degree of alternative splicing varies with species The dogmas – they are a~changing… One gene, one enzyme One gene, one polypetide One gene, one set of transcripts Alternative Splicing A very short introduction (in plants) Alternative Splicing The exons and introns of a particular gene get shuffled to create multiple isoforms of a particular protein •First demonstrated in the late 1970’s in adenovirus •Fairly well characterized in animals (at least somewhat better than in plants) •Contributes to protein diversity •Affects mRNA stability Alternative splicing in disease • By virtue of its widespread involvement in most of the genomic landscape, AS is important in almost all gene families • AS (or mis-splicing) is a very important component of genetic diseases How are AS events detected? • • • • Based on cDNA and EST data Alignment against genome sequence High-throughput RNA-seq PCR based assays Alternative splicing in metazoans Splice statistics for human genes Alternative splicing in animals. Nature Genetics Research 36; 2004 Bridging the gap between genome and transcriptome Nucleic Acids Research 32, 2004. • Alternative splicing well characterized in animals. • As many as 96% of human genes may have multiple splice forms. • Functional significance of alternative spicing still poorly understood. Alternative splicing in plants RuBisCo alternative splicing one of first plant examples: “The data presented here demonstrate the existence of alternative splicing in plant systems, but the physiological significance of synthesizing two forms of rubisco activase remains unclear. However, this process may have important implications in photosynthesis. If these polypeptides were functionally equivalent enzymes in the chloroplast, there would be no need for the production of both….” Biological significance of AS in plants …includes: - regulation of flowering; - resistance to diseases; - enzyme activity (timing, duration, turn-over time, location). Most genome databases give alternatively spliced plant gene variants Example: Disease resistance in tobacco ii - Nicotiana tabacum resistance gene N involved in resistance to TMV. Alternative splicing required to achieve resistance. Alternative transcripts Ns (short) and NL (long). NS encodes full-length, NL a truncated protein. Splicevariants produced by alternative splicing confer resistance (D). Splicevariants produced by cDNAs do not confer resistance (A, B, C). Example: Jasmonate signaling in Arabidopsis - Plant hormone; affects cell division, growth, reproduction and responses to insects, pathogens, and abiotic stress factors. - Jasmonate Signaling Repressor Protein JAZ 10 splice variants JAZ 10.1, JAZ 10.3 and JAZ 10.4 differ in susceptibility to degradation. - Phenotypic effects include male sterility, altered root growth. Example: Jasmonate signaling in Arabidopsis - Alternative splice sites C’ and D’ lead to different splice variants JAZ10.3: premature stop codon in D exon, intact JAS domain JAZ10.4: truncated C exon, protein lacks JAS domain JAZ 10 encoded by At5G13220 Sequence & course material repository http://gfx.dnalc.org/files/evidence Don’t open items, save them to your computer!! • • • • • • • Annotation (sequences & evidence) Manuals (DNA, Subway, Apollo, JalView) Presentations (.ppt files) Prospecting (sequences) Readings (Bioinformatics tools, splicing, etc.) Worksheets (Word docs, handouts, etc.) BCR-ABL (temporary; not course-related)