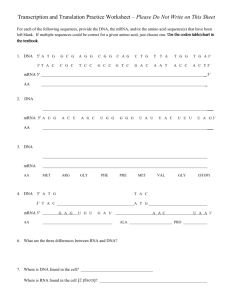
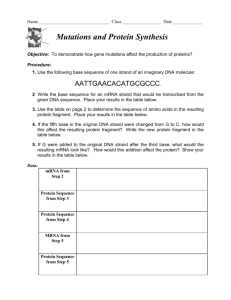
symp09_hill - University of Otago
advertisement

ENGINEERING CYANOBACTERIA FOR BIOFUEL PRODUCTION Ryan Hill, PhD candidate, Biochemistry Life is complex Metabolic engineers write “software” for living systems Living things are complex machines Bacteria are essentially self-replicating micromachines Components are at the nano-/molecular scale Life is “programmable” – Software is written in DNA and executed by the cell machinery Key Molecules and enzymes Key Molecules DNA : Deoxyribonucleic Acid. One very large molecule (3Mbp-10+Gbp). Master copy mRNA: messenger Ribonucleic Acid. Lots of small molecules (1-10kbp). Working copies Enzyme: Amino acids. Proteins that catalyze chemical reaction. Molecular machines. Key Enzymes: RNA Polymerase: Molecular machine that creates mRNA instructions from DNA templates (photocopier) Ribosome: Molecular machine that reads mRNA and builds an enzyme from the instructions encoded (robot assembler) Gene: A region of DNA that encodes all the information necessary for producing an enzyme Promoter: Region of gene that promotes transcription Terminator: Stops transcription RBS: Ribosomal binding site ORF: Open reading frame. Region of a gene that encodes the enzyme information read by the ribosome Plasmid: Small circular hoop of DNA, “Mini chromosome” encoding1-10 genes, 3-50kbp The most important process of life Transcription mRNA Promoter DNA DNA+RNA polymerase ORF RBS Terminator Protein X assembly Protein X mRNA+Ribosome Translation Ribosome Synechocystis sp. PCC 6803 Single cell bacterium (cyanobacterium) Photosynthetic – fixes carbon dioxide Genome sequenced (3.5Mbp), well understood Genome can be easily and precisely modified Butanol – Ethanols’ big brother Comparable to petroleum 91-96 fuels Compatible with current infrastructure Compatible with current vehicles/engines BUT current bio-production is inefficient Clostridium beijerinckii or C. acetobutylicum Acetone-Butanol-Ethanol (ABE) fermentation Ethanol Butanol The many uses of butanol “Until around 2005, butanol was only considered to be a bulk chemical precursor for production of acrylate and methacrylate esters, glycol ethers, butyl acetate, butylamines, and amino resins. Their use is manifold: production of adhesives/scalants, alkaloids, antibiotics, camphor, deicing fluid, dental products, detergents, elastomers, electronics, emulsifiers, eye makeup, fibers, flocculants, flotation aids (e.g., butyl xanthate), hard-surface cleaners, hormones and vitamins, hydraulic and brake fluids, industrial coatings, lipsticks, nail care products, paints, paint thinners, perfumes, pesticides, plastics, printing ink, resins, safety glass, shaving and personal hygiene products, surface coatings, super absorbents, synthetic fruit flavoring, textiles, as mobile phases in paper and thin-layer chromatography, as oil additive, as well as for leather and paper finishing” Durrie (2007) Biotechnol J 2, 1525-1534 Putting it together – Why? Synechocystis offers several advantages: Do not require a feedstock (arable land), it makes it own Grows in water There is no processing of biomass Majority of fixed carbon converted to bio-fuel The growth procedure is also a butanol extraction procedure (gas stripping) A near pure stream of butanol should be achievable directly from the bioreactor Synechocystis poly[hydroxybutyrate] (PHB) pathway PHB production is circadian controlled, i.e. turned on at night and shut down in the day Metabolic pathways Clostridium beijerinckii butanol pathway Plasmids Two base plasmids: pRH-ECT7 – Knock-out of phaEC, inserts ORFs under control the phaEC promoter/RBS, uses T7 terminator pRH-BT7b – Knock-in extra ORF(s) onto the end of the phaAB mRNA (after phaB), has RBS from psbA2 gene, uses T7 terminator Expression plasmids – ECT7 pRH-ECT7::luxAB, kan Synechocystis strains pRH-ECT7 based: ∆phaEC::aph (kanamycin resistance) ∆phaEC::luxAB, aph pRH-BT7b based: ∆phaAB::cat (chloramphenicol resistance) ∆phaAB::luxAB, cat PHB Detection Wildtype ∆phaAB::cat ∆phaEC::aph ∆phaAB::luxAB, cat ∆phaEC::luxAB, aph Testing the programs – ECT7::luxAB Normalised luciferase activity from ∆phaEC::luxAB, aph 110.0 60 100.0 50 90.0 80.0 60.0 30 50.0 40.0 20 30.0 20.0 10 10.0 0.0 0 0 2 4 6 8 10 12 Time, hours 14 16 18 20 22 24 Light, umol.m-2.s-1 Activity, % maximum 40 70.0 Testing the programs – BT7b::luxAB Normalised luciferase activity from ∆phaAB::luxAB, cat 110 60 100 50 90 80 Activity, % maximum 40 70 60 30 50 40 20 30 20 10 10 0 0 0 2 4 6 Time, hours 8 10 12 Summary Plasmids pRH-ETC7 and –BT7b constructed Modification of PHB metabolism doesn’t damage Synechocystsis pRH-ECT7 successfully knocks out PHB production pRH-BT7b successfully does not damage PHB production Both pRH-ECT7 and –BT7b work as intended when using luciferase as a reporter Acknowledgements Department of Biochemistry Assoc. Prof. Julian Eaton-Rye Assoc. Prof. John Cutfield Funding -Department of Biochemistry -OERC -University of Otago