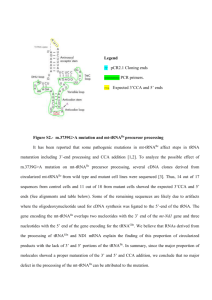
2.1. Selection of aptamers from computational results
advertisement

SUPPLEMENTARY MATERIAL Computational selection of RNA aptamer against angiopoietin-2 and experimental evaluation Wen-Pin Hua,b*, Jangam Vikram Kumara*, Chun-Jen Huangc, Wen-Yih Chend a b Department of Biomedical Informatics, Asia University, Taichung City, Taiwan Department of Medical Laboratory Science and Biotechnology, China Medical University, Taichung City, Taiwan c Graduate Institute of Biomedical Engineering, National Central University, Jhong-Li City, Taiwan d Department of Chemical and Materials Engineering, National Central University, Jhong-Li City, Taiwan Corresponding author: Wen-Pin Hu; Jangam Vikram Kumar Department of Biomedical Informatics, Asia University 500, Lioufeng Rd., Wufeng, Taichung 41354, Taiwan TEL: +886-4-23323456 ext.20022; FAX: +886-4-23305737 E-mail: wenpinhu@asia.edu.tw (W.P. Hu); vikramkumar.bioinf@gmail.com (J.V. Kumar) 1. Materials and methods 1.1 Computational assay Figure S1.Model of the 3D Structure of Seq15. This model is generated by using the CentroidFold and RNAComposer webservers. 1.2 The library of mutated RNA sequences Table S1.Nucleotides of three RNA aptamers involved in the binding interface with Ang2. Nucleotides at position 10 to 16 and 33 to 99 were marked with bold, underlined text. Name Nucleotides involved in binding interface with Ang2 Seq1 A4 G5 C6 C7 U8 C9 A10 A13 G14 C15 U16 C17 A18 G30 C31 C32 U33 G34 G35 A36 Seq2 U2 A3 A4 C5 C6 A7 U8 C9 A10 G11 C12 U13 C14 A15 U16A39 C40 Seq3 C28 U29 C30 A31 C32 C33 C34 A35 C36 A37 C38 C39 G40 Seq4 C1 C2 A3 C4 C5 G6 A7 U8 C9 G10 C11 A12 U13 C14 A15 G16 Seq5 U9 C10 U11 G18 C19 C20 G21 A22 U23 C24 A25 U26 C27 G29 C30 G31 C32 U33 Seq6 A5 C6 C7 A8 C9 G10 C11 U14 A15 U16 C17 A18 G19 C20 U21 A22 A23 C34 U35 A36 Seq7 C6 C7 A8 G9 U10 C11 A12 C13 C14 A15 U16 Seq8 C4 C5 A6 A7 G8 C10 U11 C12 A13 C14 G15 U16 U17 Seq9 G1 G2 A3 G4 C5 G6 C7 A8 A9 G25 A26 A27 C28 U29 C30 C31 G32 Seq10 C13 U14 U15 A16 G29 A30 A31 C32 U33 C34 C35 A36 G37 A38 Table S1. (Continued) Nucleotides of three RNA aptamers involved in binding interface with Ang2. Nucleotides at position 10 to 16 and 33 to 99 were marked with bold, underlined text. Seq11 A26 A27 C28 U29 C30 C31 A32 U33 G34 C35 A36 Seq12 G23 U24 U25 G26 G28 U29 C30 U31 C32 G33 U34 C35 G36 A37 A38 Seq13 C1 A2 C3 U4 C5 A6 G7 C8 G9 C10 C11 C12 U13 G14 C15 G16 A17 A18 C32 C39 U40 Seq14 G22 A23 A24 C25 U26 C27 C28 U29 G30 C32 C33 C34 U35 C36 U37 A38 C39 Seq15 G10 C11 G12 G13 A14 C20 U21 C22 A23 U24 C37 C38 Seq16 C13 C14 A15 C16 U17 C20 C21 A22 A23 C24 C25 U27 1.3 SPR imaging apparatus The SPR platform is a self- referencing SPR imaging sensor with polarization contrast, which is able to detect the smallest signal corresponding to the change in the refractive index unit (RIU) better than10-6 within the operation range of 0.011 RIU. The p-polarized narrow-band light beam (central emission wavelength is 750 nm) impinges on a glass substrate and excites SPs at the metal-dielectric interface. It also can be illustrated in terms of protein surface coverage that the sensor can detect the changes that are as small as 0.2 pg/mm2. The SPRi can be utilized by conducting traditional measurements or performing a high-throughput analysis with the use of an array-formatted chip. The SPR chips were prepared by using clean BK7 glass substrates. The glass substrates were pre-coated with an adhesion layer of chromium (thickness approx. 2 nm), and an active gold layer with a thickness of 48 nm was subsequently coated via an evaporation deposition process at pressures below 110-6 Torr. All experiments were performed at a constant temperature of 25°C and at a flow rate of 50 μl/min. Six flow channels were used in the SPR experiments, and one channel was used as a reference channel. 2. Results and discussion 2.1. Selection of aptamers from computational results Mutant sequence of Seq1 Table S2.The simulation results of 189 mutant RNA aptamer sequences. The mutated positions in the sequences and the ZRNAK scores of three selected aptamers were marked with bold letters. ZRANK Type Sequence(5-3) Score ACUAGCCUCUUCAGCUCAUGUGCCCCUCCGCCAGGAUCAC -66.174 ACUAGCCUCUUCAGCUCAUGUGCCCCUCCGCCGGGAUCAC -73.649 ACUAGCCUCUUCAGCUCAUGUGCCCCUCCGCCCGGAUCAC -69.671 ACUAGCCUCGUCAGCUCAUGUGCCCCUCCGCCAGGAUCAC -63.625 ACUAGCCUCGUCAGCUCAUGUGCCCCUCCGCCGGGAUCAC -61.696 ACUAGCCUCGUCAGCUCAUGUGCCCCUCCGCCCGGAUCAC -71.018 ACUAGCCUCCUCAGCUCAUGUGCCCCUCCGCCAGGAUCAC -74.2 ACUAGCCUCCUCAGCUCAUGUGCCCCUCCGCCGGGAUCAC -62.316 ACUAGCCUCCUCAGCUCAUGUGCCCCUCCGCCCGGAUCAC -66.705 ACUAGCCUCAACAGCUCAUGUGCCCCUCCGCCUAGAUCAC -73.514 ACUAGCCUCAACAGCUCAUGUGCCCCUCCGCCUCGAUCAC -74.701 ACUAGCCUCAACAGCUCAUGUGCCCCUCCGCCUUGAUCAC -87.666 ACUAGCCUCAGCAGCUCAUGUGCCCCUCCGCCUAGAUCAC -66.493 ACUAGCCUCAGCAGCUCAUGUGCCCCUCCGCCUCGAUCAC -66.839 ACUAGCCUCAGCAGCUCAUGUGCCCCUCCGCCUUGAUCAC -61.193 ACUAGCCUCACCAGCUCAUGUGCCCCUCCGCCUAGAUCAC -87.829 ACUAGCCUCACCAGCUCAUGUGCCCCUCCGCCUCGAUCAC -74.337 ACUAGCCUCACCAGCUCAUGUGCCCCUCCGCCUUGAUCAC -65.241 ACUAGCCUCAUAAGCUCAUGUGCCCCUCCGCCUGAAUCAC -71.17 ACUAGCCUCAUAAGCUCAUGUGCCCCUCCGCCUGCAUCAC -76.63 ACUAGCCUCAUAAGCUCAUGUGCCCCUCCGCCUGUAUCAC -68.573 ACUAGCCUCAUGAGCUCAUGUGCCCCUCCGCCUGAAUCAC -74.78 ACUAGCCUCAUGAGCUCAUGUGCCCCUCCGCCUGCAUCAC -72.671 ACUAGCCUCAUGAGCUCAUGUGCCCCUCCGCCUGUAUCAC -73.072 ACUAGCCUCAUUAGCUCAUGUGCCCCUCCGCCUGAAUCAC -68.47 ACUAGCCUCAUUAGCUCAUGUGCCCCUCCGCCUGCAUCAC -83.747 ACUAGCCUCAUUAGCUCAUGUGCCCCUCCGCCUGUAUCAC -64.297 ACUAGCCUCAUCUGCUCAUGUGCCCCUCCGCCUGGCUCAC -77.279 ACUAGCCUCAUCUGCUCAUGUGCCCCUCCGCCUGGGUCAC -70.501 ACUAGCCUCAUCUGCUCAUGUGCCCCUCCGCCUGGUUCAC -73.393 Mutant sequence of Seq1 Table S2.(Continued) The simulation results of 189 mutant RNA aptamer sequences. The mutated positions in the sequences and the ZRNAK scores of three selected aptamers were marked with bold letters. ZRANK Type Sequence(5-3) Score ACUAGCCUCAUCGGCUCAUGUGCCCCUCCGCCUGGCUCAC -69.012 ACUAGCCUCAUCGGCUCAUGUGCCCCUCCGCCUGGGUCAC -60.148 ACUAGCCUCAUCGGCUCAUGUGCCCCUCCGCCUGGUUCAC -71.464 ACUAGCCUCAUCCGCUCAUGUGCCCCUCCGCCUGGCUCAC -79.954 ACUAGCCUCAUCCGCUCAUGUGCCCCUCCGCCUGGGUCAC -76.995 ACUAGCCUCAUCCGCUCAUGUGCCCCUCCGCCUGGUUCAC -78.571 ACUAGCCUCAUCAACUCAUGUGCCCCUCCGCCUGGAACAC -77.097 ACUAGCCUCAUCAACUCAUGUGCCCCUCCGCCUGGACCAC -67.97 ACUAGCCUCAUCAACUCAUGUGCCCCUCCGCCUGGAGCAC -74.329 ACUAGCCUCAUCACCUCAUGUGCCCCUCCGCCUGGAACAC -65.604 ACUAGCCUCAUCACCUCAUGUGCCCCUCCGCCUGGACCAC -70.131 ACUAGCCUCAUCACCUCAUGUGCCCCUCCGCCUGGAGCAC -73.143 ACUAGCCUCAUCAUCUCAUGUGCCCCUCCGCCUGGAACAC -72.094 ACUAGCCUCAUCAUCUCAUGUGCCCCUCCGCCUGGACCAC -66.34 ACUAGCCUCAUCAUCUCAUGUGCCCCUCCGCCUGGAGCAC -78.013 ACUAGCCUCAUCAGAUCAUGUGCCCCUCCGCCUGGAUAAC -69.839 ACUAGCCUCAUCAGAUCAUGUGCCCCUCCGCCUGGAUGAC -67.988 ACUAGCCUCAUCAGAUCAUGUGCCCCUCCGCCUGGAUUAC -78.518 ACUAGCCUCAUCAGGUCAUGUGCCCCUCCGCCUGGAUAAC -66.59 ACUAGCCUCAUCAGGUCAUGUGCCCCUCCGCCUGGAUGAC -68.432 ACUAGCCUCAUCAGGUCAUGUGCCCCUCCGCCUGGAUUAC -77.178 ACUAGCCUCAUCAGUUCAUGUGCCCCUCCGCCUGGAUAAC -66.492 ACUAGCCUCAUCAGUUCAUGUGCCCCUCCGCCUGGAUGAC -61.954 ACUAGCCUCAUCAGUUCAUGUGCCCCUCCGCCUGGAUUAC -66.717 ACUAGCCUCAUCAGCACAUGUGCCCCUCCGCCUGGAUCCC -84.072 ACUAGCCUCAUCAGCACAUGUGCCCCUCCGCCUGGAUCGC -67.44 ACUAGCCUCAUCAGCACAUGUGCCCCUCCGCCUGGAUCUC -67.844 ACUAGCCUCAUCAGCGCAUGUGCCCCUCCGCCUGGAUCCC -68.77 ACUAGCCUCAUCAGCGCAUGUGCCCCUCCGCCUGGAUCGC -64.424 ACUAGCCUCAUCAGCGCAUGUGCCCCUCCGCCUGGAUCUC -65.86 ACUAGCCUCAUCAGCCCAUGUGCCCCUCCGCCUGGAUCCC -65.771 ACUAGCCUCAUCAGCCCAUGUGCCCCUCCGCCUGGAUCGC -77.968 ACUAGCCUCAUCAGCCCAUGUGCCCCUCCGCCUGGAUCUC -74.137 Mutant sequence of Seq2 Table S2.(Continued) The simulation results of 189 mutant RNA aptamer sequences. The mutated positions in the sequences and the ZRNAK scores of three selected aptamers were marked with bold letters. ZRANK Type Sequence(5-3) Score UUAACCAUCUGCUCAUGGCCCCUGCCCUCUCAUGGACCAC -87.021 UUAACCAUCUGCUCAUGGCCCCUGCCCUCUCAGGGACCAC -78.846 UUAACCAUCUGCUCAUGGCCCCUGCCCUCUCACGGACCAC -73.147 UUAACCAUCGGCUCAUGGCCCCUGCCCUCUCAUGGACCAC -61.103 UUAACCAUCGGCUCAUGGCCCCUGCCCUCUCAGGGACCAC -71.789 UUAACCAUCGGCUCAUGGCCCCUGCCCUCUCACGGACCAC -73.519 UUAACCAUCCGCUCAUGGCCCCUGCCCUCUCAUGGACCAC -68.534 UUAACCAUCCGCUCAUGGCCCCUGCCCUCUCAGGGACCAC -67.404 UUAACCAUCCGCUCAUGGCCCCUGCCCUCUCACGGACCAC -71.113 UUAACCAUCAACUCAUGGCCCCUGCCCUCUCAAAGACCAC -83.898 UUAACCAUCAACUCAUGGCCCCUGCCCUCUCAAUGACCAC -70.853 UUAACCAUCAACUCAUGGCCCCUGCCCUCUCAACGACCAC -68.273 UUAACCAUCAUCUCAUGGCCCCUGCCCUCUCAAAGACCAC -87.682 UUAACCAUCAUCUCAUGGCCCCUGCCCUCUCAAUGACCAC -63.614 UUAACCAUCAUCUCAUGGCCCCUGCCCUCUCAACGACCAC -66.266 UUAACCAUCACCUCAUGGCCCCUGCCCUCUCAAAGACCAC -72.281 UUAACCAUCACCUCAUGGCCCCUGCCCUCUCAAUGACCAC -76.843 UUAACCAUCACCUCAUGGCCCCUGCCCUCUCAACGACCAC -77.119 UUAACCAUCAGAUCAUGGCCCCUGCCCUCUCAAGAACCAC -82.028 UUAACCAUCAGAUCAUGGCCCCUGCCCUCUCAAGUACCAC -58.122 UUAACCAUCAGAUCAUGGCCCCUGCCCUCUCAAGCACCAC -97.609 UUAACCAUCAGUUCAUGGCCCCUGCCCUCUCAAGAACCAC -69.531 UUAACCAUCAGUUCAUGGCCCCUGCCCUCUCAAGUACCAC -70.08 UUAACCAUCAGUUCAUGGCCCCUGCCCUCUCAAGCACCAC -78.163 UUAACCAUCAGGUCAUGGCCCCUGCCCUCUCAAGAACCAC -87.09 UUAACCAUCAGGUCAUGGCCCCUGCCCUCUCAAGUACCAC -84.423 UUAACCAUCAGGUCAUGGCCCCUGCCCUCUCAAGCACCAC -67.203 UUAACCAUCAGCACAUGGCCCCUGCCCUCUCAAGGUCCAC -69.167 UUAACCAUCAGCACAUGGCCCCUGCCCUCUCAAGGGCCAC -70.366 UUAACCAUCAGCACAUGGCCCCUGCCCUCUCAAGGCCCAC -73.611 UUAACCAUCAGCGCAUGGCCCCUGCCCUCUCAAGGUCCAC -74.263 UUAACCAUCAGCGCAUGGCCCCUGCCCUCUCAAGGGCCAC -79.564 Mutant sequence of Seq2 Table S2.(Continued) The simulation results of 189 mutant RNA aptamer sequences. The mutated positions in the sequences and the ZRNAK scores of three selected aptamers were marked with bold letters. ZRANK Type Sequence(5-3) Score UUAACCAUCAGCGCAUGGCCCCUGCCCUCUCAAGGCCCAC -79.122 UUAACCAUCAGCCCAUGGCCCCUGCCCUCUCAAGGUCCAC -69.499 UUAACCAUCAGCCCAUGGCCCCUGCCCUCUCAAGGGCCAC -74.227 UUAACCAUCAGCCCAUGGCCCCUGCCCUCUCAAGGCCCAC -68.816 UUAACCAUCAGCUAAUGGCCCCUGCCCUCUCAAGGAACAC -68.767 UUAACCAUCAGCUAAUGGCCCCUGCCCUCUCAAGGAUCAC -77.011 UUAACCAUCAGCUAAUGGCCCCUGCCCUCUCAAGGAGCAC -64.889 UUAACCAUCAGCUUAUGGCCCCUGCCCUCUCAAGGAACAC -79.081 UUAACCAUCAGCUUAUGGCCCCUGCCCUCUCAAGGAUCAC -84.05 UUAACCAUCAGCUUAUGGCCCCUGCCCUCUCAAGGAGCAC -71.727 UUAACCAUCAGCUGAUGGCCCCUGCCCUCUCAAGGAACAC -77.511 UUAACCAUCAGCUGAUGGCCCCUGCCCUCUCAAGGAUCAC -67.982 UUAACCAUCAGCUGAUGGCCCCUGCCCUCUCAAGGAGCAC -72.09 UUAACCAUCAGCUCUUGGCCCCUGCCCUCUCAAGGACUAC -69.207 UUAACCAUCAGCUCUUGGCCCCUGCCCUCUCAAGGACGAC -72.46 UUAACCAUCAGCUCGUGGCCCCUGCCCUCUCAAGGACAAC -70.948 UUAACCAUCAGCUCGUGGCCCCUGCCCUCUCAAGGACUAC -72.441 UUAACCAUCAGCUCGUGGCCCCUGCCCUCUCAAGGACGAC -67.28 UUAACCAUCAGCUCCUGGCCCCUGCCCUCUCAAGGACAAC -67.681 UUAACCAUCAGCUCCUGGCCCCUGCCCUCUCAAGGACUAC -66.611 UUAACCAUCAGCUCCUGGCCCCUGCCCUCUCAAGGACGAC -78.931 UUAACCAUCAGCUCAAGGCCCCUGCCCUCUCAAGGACCUC -68.63 UUAACCAUCAGCUCAAGGCCCCUGCCCUCUCAAGGACCGC -63.629 UUAACCAUCAGCUCAAGGCCCCUGCCCUCUCAAGGACCCC -66.074 UUAACCAUCAGCUCAGGGCCCCUGCCCUCUCAAGGACCUC -86.223 UUAACCAUCAGCUCAGGGCCCCUGCCCUCUCAAGGACCGC -73.089 UUAACCAUCAGCUCAGGGCCCCUGCCCUCUCAAGGACCCC -67.327 UUAACCAUCAGCUCACGGCCCCUGCCCUCUCAAGGACCUC -66.499 UUAACCAUCAGCUCACGGCCCCUGCCCUCUCAAGGACCGC -72.857 UUAACCAUCAGCUCACGGCCCCUGCCCUCUCAAGGACCCC -60.727 Mutant sequence of Seq15 Table S2.(Continued) The simulation results of 189 mutant RNA aptamer sequences. The mutated positions in the sequences and the ZRNAK scores of three selected aptamers were marked with bold letters. ZRANK Type Sequence(5-3) Score GAGGACGAUACGGACUAGCCUCAUCAGCUCAUAUGCCCCUC -72.04 GAGGACGAUACGGACUAGCCUCAUCAGCUCAUUUGCCCCUC -71.647 GAGGACGAUACGGACUAGCCUCAUCAGCUCAUCUGCCCCUC -78.706 GAGGACGAUUCGGACUAGCCUCAUCAGCUCAUAUGCCCCUC -68.452 GAGGACGAUUCGGACUAGCCUCAUCAGCUCAUUUGCCCCUC -88.897 GAGGACGAUUCGGACUAGCCUCAUCAGCUCAUCUGCCCCUC -71.47 GAGGACGAUCCGGACUAGCCUCAUCAGCUCAUAUGCCCCUC -63.921 GAGGACGAUCCGGACUAGCCUCAUCAGCUCAUUUGCCCCUC -62.649 GAGGACGAUCCGGACUAGCCUCAUCAGCUCAUCUGCCCCUC -76.687 GAGGACGAUGAGGACUAGCCUCAUCAGCUCAUGAGCCCCUC -88.326 GAGGACGAUGAGGACUAGCCUCAUCAGCUCAUGGGCCCCUC -79.479 GAGGACGAUGAGGACUAGCCUCAUCAGCUCAUGCGCCCCUC -76.345 GAGGACGAUGGGGACUAGCCUCAUCAGCUCAUGAGCCCCUC -86.981 GAGGACGAUGGGGACUAGCCUCAUCAGCUCAUGGGCCCCUC -76.127 GAGGACGAUGGGGACUAGCCUCAUCAGCUCAUGCGCCCCUC -76.427 GAGGACGAUGUGGACUAGCCUCAUCAGCUCAUGAGCCCCUC -75.317 GAGGACGAUGUGGACUAGCCUCAUCAGCUCAUGGGCCCCUC -76.279 GAGGACGAUGUGGACUAGCCUCAUCAGCUCAUGCGCCCCUC -73.258 GAGGACGAUGCAGACUAGCCUCAUCAGCUCAUGUACCCCUC -78.506 GAGGACGAUGCAGACUAGCCUCAUCAGCUCAUGUUCCCCUC -81.902 GAGGACGAUGCAGACUAGCCUCAUCAGCUCAUGUCCCCCUC -66.779 GAGGACGAUGCUGACUAGCCUCAUCAGCUCAUGUACCCCUC -86.916 GAGGACGAUGCUGACUAGCCUCAUCAGCUCAUGUUCCCCUC -89.858 GAGGACGAUGCUGACUAGCCUCAUCAGCUCAUGUCCCCCUC -76.02 GAGGACGAUGCCGACUAGCCUCAUCAGCUCAUGUACCCCUC -70.982 GAGGACGAUGCCGACUAGCCUCAUCAGCUCAUGUUCCCCUC -72.649 GAGGACGAUGCCGACUAGCCUCAUCAGCUCAUGUCCCCCUC -93.335 GAGGACGAUGCGAACUAGCCUCAUCAGCUCAUGUGACCCUC -81.726 GAGGACGAUGCGAACUAGCCUCAUCAGCUCAUGUGUCCCUC -81.845 GAGGACGAUGCGAACUAGCCUCAUCAGCUCAUGUGGCCCUC -77.127 GAGGACGAUGCGUACUAGCCUCAUCAGCUCAUGUGACCCUC -79.628 GAGGACGAUGCGUACUAGCCUCAUCAGCUCAUGUGUCCCUC -86.623 GAGGACGAUGCGUACUAGCCUCAUCAGCUCAUGUGGCCCUC -79.604 Mutant sequence of Seq15 Table S2.(Continued) The simulation results of 189 mutant RNA aptamer sequences. The mutated positions in the sequences and the ZRNAK scores of three selected aptamers were marked with bold letters. ZRANK Type Sequence(5-3) Score GAGGACGAUGCGCACUAGCCUCAUCAGCUCAUGUGACCCUC -72.09 GAGGACGAUGCGCACUAGCCUCAUCAGCUCAUGUGUCCCUC -67.432 GAGGACGAUGCGCACUAGCCUCAUCAGCUCAUGUGGCCCUC -87.531 GAGGACGAUGCGGUCUAGCCUCAUCAGCUCAUGUGCACCUC -67.705 GAGGACGAUGCGGUCUAGCCUCAUCAGCUCAUGUGCUCCUC -59.599 GAGGACGAUGCGGUCUAGCCUCAUCAGCUCAUGUGCGCCUC -66.643 GAGGACGAUGCGGGCUAGCCUCAUCAGCUCAUGUGCACCUC -88.951 GAGGACGAUGCGGGCUAGCCUCAUCAGCUCAUGUGCUCCUC -78.324 GAGGACGAUGCGGGCUAGCCUCAUCAGCUCAUGUGCGCCUC -87.948 GAGGACGAUGCGGCCUAGCCUCAUCAGCUCAUGUGCACCUC -67.698 GAGGACGAUGCGGCCUAGCCUCAUCAGCUCAUGUGCUCCUC -72.326 GAGGACGAUGCGGCCUAGCCUCAUCAGCUCAUGUGCGCCUC -74.909 GAGGACGAUGCGGAAUAGCCUCAUCAGCUCAUGUGCCACUC -81.962 GAGGACGAUGCGGAAUAGCCUCAUCAGCUCAUGUGCCUCUC -78.683 GAGGACGAUGCGGAAUAGCCUCAUCAGCUCAUGUGCCGCUC -74.477 GAGGACGAUGCGGAUUAGCCUCAUCAGCUCAUGUGCCACUC -75.669 GAGGACGAUGCGGAUUAGCCUCAUCAGCUCAUGUGCCUCUC -72.017 GAGGACGAUGCGGAUUAGCCUCAUCAGCUCAUGUGCCGCUC -89.904 GAGGACGAUGCGGAGUAGCCUCAUCAGCUCAUGUGCCACUC -77.132 GAGGACGAUGCGGAGUAGCCUCAUCAGCUCAUGUGCCUCUC -75.5 GAGGACGAUGCGGAGUAGCCUCAUCAGCUCAUGUGCCGCUC -85.028 GAGGACGAUGCGGACAAGCCUCAUCAGCUCAUGUGCCCAUC -78.824 GAGGACGAUGCGGACAAGCCUCAUCAGCUCAUGUGCCCUUC -67.014 GAGGACGAUGCGGACAAGCCUCAUCAGCUCAUGUGCCCGUC -70.113 GAGGACGAUGCGGACGAGCCUCAUCAGCUCAUGUGCCCAUC -77.709 GAGGACGAUGCGGACGAGCCUCAUCAGCUCAUGUGCCCUUC -85.681 GAGGACGAUGCGGACGAGCCUCAUCAGCUCAUGUGCCCGUC -72.912 GAGGACGAUGCGGACCAGCCUCAUCAGCUCAUGUGCCCAUC -79.488 GAGGACGAUGCGGACCAGCCUCAUCAGCUCAUGUGCCCUUC -76.647 GAGGACGAUGCGGACCAGCCUCAUCAGCUCAUGUGCCCGUC -88.449