Energy Metabolism
advertisement

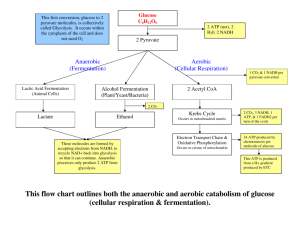
Energy Metabolism • ATP synthesis – Outline the steps of glycolysis – Outline the steps of lipolysis – Citric acid cycle/Electron transport chain • Control processes – Explain the contribution of mass action to the rate of ATP synthesis – Similarly, allosteric feedback Phospho-creatine ATP buffer • Creatine Kinase – Unique to striated muscle – Creatine + ATP ADP + phospho-creatine • Creatine – 20-40 mM total creatine – 16-32 mM phospho – ATP ~ 5-10 mM Glycolysis • Convert Glucose to Pyruvate – Yield 2 ATP + 2 NADH per glucose – Consume 2 ATP to form 2x glyceraldehyde phosphate – Produce 2 ATP + 1 NADH per GAP • Carefully controlled – 12 different enzyme-catalyzed steps – Limited by phosphofructokinase – Limited by substrate availability Glycolysis: phosphorylation • ATP consuming – Glucose phosphorylation by hexokinase – Fructose phosphorylation by phosphofructokinase • Triose phosphate isomerase Glycolysis: oxidation • Pyruvate kinase – Transfer Pi to ADP – Driven by oxidative potential of 2’ O • Summary – Start C6H12O6 – End 2xC3H3O3 – Added 0xO – Lost 6xH – Gained 2xNADH, 2xATP GAPDH NADH ATP phosphoglycerate kinase pyruvate kinase Pyruvate • Lactic Acid – Regenerates NAD+ – Redox neutral • Ethanol – Regenerates NAD+ – Redox neutral • Acetyl-CoA – Pyruvate import to mitocondria – ~15 more ATP per pyruvate pyruvate 2-HydroxyethylThiamine diphosphate S-acetyldihydrolipoyllysine Acetyl-CoA Carbohydrate metabolism depends on transport • H+, pyruvate cotransporter Major Facilitator Superfamily Monocarboxylate transporter Competition between H+ driven transport to mitochondria and NADH/H+ driven conversion to lactate Cytoplasmic NADH is also used to generate mitochondrial FADH2, coupling transport to ETC saturation “glycerol-3P shuttle” Halestrap & Price 1999 Gluconeogenesis • During contraction, inefficient glycolysis wastes glucose – Many glycolytic enzymes are reversible • Special enzymes – Pyruvate carboxylase – Phosphoenyl pyruvate carboxykinase • Swap carboxyl group for phosphate • Generates 3-C phosphoenolpyruvate from OA – Fructose-1,6-bisphosphatase • Generates fructose-6-phosphate Mitochondrial • Generate 4-C oxaloacetate from 3-C pyruvate Fatty Acid/b-oxidation Cycle 1x FADH2 1x NADH Acetyl-CoA – 3x NADH+ –1xFADH2 • Acyl(n)-CoA + NAD+ + FAD Acyl(n-2)-CoA + Acetyl-CoA + NADH +FADH2 Carnitine palmitoyltransferase Fatty acid elongation Acyl-CoA Acyl-CoA synthase FAD Acyl-CoA dehydrogenase Acyl-CoA FADH2 acetyl-CoA acyltransferase Acetyl-CoA Didehydroacyl-CoA Acyl-CoA hydrase CoA-SH 3-hydroxyacyl-CoA dehydrogenase Hydroxyacyl-CoA Oxoacyl-CoA NADH NAD+ Reactive oxygen • FADH2 oxidative stress – Succinate; saturated FA – FADH2 + Fe3+ FADH • + H+ + Fe2+ – Fe2+ + H2O2Fe3+ + OH- + OH• • FADH2 more completely reduces UQ than does NADH Acyl-CoA Acyl-CoA FAD Acyl-CoA dehydrogenase UQ ETF:QO oxidoreductase FADH2 Didehydroacyl-CoA FADH2 FAD O2 Acyl-CoA oxidase UQH2 H2O2 Didehydroacyl-CoA Free fatty acids from triglycerides • FFA cleavage from circulating lipoproteins – Protein/cholesterol carriers: Lipoprotein • Density inversely correlates with lipid • Correlates with cholesterol/FA (except HDL) • VLDL & LDL to IDL – Lipoprotein lipase (LPL) – HDL scavenges cholesterol & facilitates IDL breakdown • Triglycerides are retained in intracellular droplets – Don’t fit in membrane (no phosphate) – Not water soluble Fatty acid metabolism depends on transport • FAAcyl-CoA Acyl-Carnitine Acyl-CoA Cytoplasm Intermembrane Matrix Working substrate Boron & Boulpaep Mitochondrial Transport • • • • • Carrier protein (FABP) Long chain acyl-CoA synthetase (LCAS) Cross outer membrane via porin Convert to acylcarnitine in intermembrane Cross inner membrane via carnitine:acylcarnitine transferase • Convert back to acyl-CoA in matrix Mitochondrial Structure • Principal metabolic engine • Symbiotic bacteria – 6k-370kBP genome – Human: 13 proteins • Dual membrane – ie: two bilayers – Outer membrane highly permeable – Inner membrane highly impermeable Mitochondrial Matrix • Highly oxidative environment • Unique proton gradient – High pH (8), negative (-180 mV), ~18 kJ/mole – H+ actively transported out of matrix – H+ leak back as H+PO4 2- • Capture gradient energy for ATP synthesis – H+ ATPase pump – ADP-ATP antiporter • Other proton co-transporters – Pyruvate, citrate – Glutamate, citruline Metabolic Substrates • Sugars – Metabolized in cytoplasm to pyruvate – Co-transported to matrix with H+ – Bound to Coenzyme A as Acetyl-CoA • Fatty acids – To intermembrane space as Acyl-CoA – To matrix as Acyl-carnitine – Metabolized to Acetyl-CoA in matrix • Proteins CH3 C=O COO- Acetyl Coenzyme A • Common substrate for oxidative metabolism • S-linked acetate carrier The Citric Acid Cycle Acetyl-Coenzyme A CoA These carbons will be removed NADH Oxaloacetate New carbons Citrate Carbon Oxygen Malate Coenzyme A Isocitrate NADH + = Fumarate a-Ketoglutarate FADH2 Succinate CoA Succinyl CoA + GTP NADH CoA Electron transport • Couple NADH/FADH2 electrons to H+ export – Ideally this completes NADH + H+ + ½ O2 NAD+ + H++2e½O2+2 H++ 2e- NADH DE0=-0.32V H2O DE0=0.82V – Electron leakage NAD+ +H2O KEGG pathway Enzyme Commission (EC) number •Hierarchical •Function-centric nomenclature •Compare •Gene Ontology (GO) ID •Entrez RefSeq •UniProt ID Metabolite KEGG http://www.genome.jp/kegg/pathway.html Cyclic redox reactions Oxidized NAD+ FAD NADH CoQ/ubiquinone Cyto-C3+ O2 dihydroubiquinone Cyto-C2+ H 2O FADH2 Reduced You can only have this progressive redox process if molecular position is carefully controlled NAD+ NADH FAD FADH2 Ubuquinone Cytochrome C O2 H2O E0 = -0.32V E0 = -0.22V E0 = 0.10V E0 = 0.22V E0 = 0.82V Proton ATPase/Complex V • ATP driven proton pump – “Reversible” – Couples H+ gradient to ATP synthesis Fatty acid/carbohydrate oxidation • Oxygen – CnH2n + 3/2 n O2 n CO2+ n H2O – CnH2nOn +n O2 n CO2 + n H2O – Respiratory Quotient CO2/O2 • 0.67 Fatty acids • 1.00 Carbohydrates • Adenine electron transporters – 6-C glucose6 NADH + 2 FADH2 (3:1) – 16-C FA 32 NADH + 16 FADH2 (2:1) • Redox chemistry differs for FA/CHO Muscle substrate utilization • Rest: fatty acids • Active: glycolysis • Recovery: – Pyruvate oxidation – Gluconeogenesis Role of mass action in flux control • Diffusion – J = D ∂f/∂x (greater flux down a steeper gradient) – ∂f/ ∂t= ∂J/∂x • Kinetics – d[P]/dt = k[S] (1st order) – d[P]/dt = Vmax [S]/(Km + [S]) (Michaelis-Menten) – d[P]/dt = k [S1][S2] (2nd order) Mass action in glycolysis • Diffusion – Substrate consumption increases gradient – Increased gradient accelerates mass flow • Kinetics – G+ATPG6p d[G6p]dt = k1[G][ATP]≈k[G] – G6pF6p d[F6p]/dt = k2[G6p] – F6p+ATPF1,6p <etc> – F1,6pG3p+DAp – DApG3p Mitochondrial substrate dependence • More ADPfaster ATP – Discharge proton gradient – Lower ETC resitsance • More NADfaster – Faster NADH – Greater ETC input Wu &al 2007 Role of allosteric regulation • Allosteric – Binding to other-than-active site changes enzyme Allosteric ADP binding site kinetics – Vmax or kM • Many metabolic enzymes are regulated by downstream products – Phosphofructokinase • Citrate inhibits • ADP activates – Gylcogen synthase Active site PDB:3PFK G6P regulation of GS • Allosteric conformational change Without G6P Less active With G6P More active Baskaran et al. 2010 Role of post-translational regulation • Chemical modification of enzymes alters activity – Phosphorylation – Ribosylation, acylation, SUMOylation, etc – Integrative response to complex conditions • Insulin – Insulin IRPI3KGLUT4 translocation glucose uptake – PI3KPKB--|GSK--|GS Phospho-regulation of glycogen • PKA • PKB +GP via phosphorylase kinase -GS -PP1 via G-subunit +GS via GSK +PP1 via G-subunit •PP1 +GS -GP Activates Inhibits PK PKA GP PP1-G PP1 GS GP Glycogen Synthesis PP1 PP1-G GS GSK3 PKB AMP kinase • Allosterically activated by AMP – Adenylate kinase: 2 ADP AMP + ATP – ADP levels insensitive to energy state PFKglycolysis --|GSGlyconeogenesis --|ACCMalonyl CoA--|CPTFA oxidation --|ACClipogenesis TSC2--|mTOR…protein synthesis --|HMGCoAcholesterol synthesis Summary • Sources of ATP – Creatine – Gylcolysis: GG3p2OPA – Lipolysis: acyl-CoAoxoacyl-CoA – Citric Acid Cycle/Electron Transport Chain • AcCoACitrate...Oxaloacetate • Rate control by – Mass action – Allosteric feedback – Hormonal control