Chapter 10
advertisement

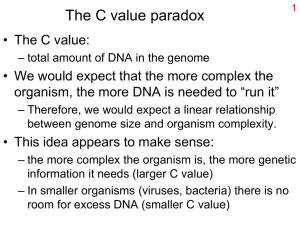
Chapter 10 Classification of Microorganisms Terminology • Taxonomy – The science of classifying organisms – Provides universal names for organisms – Provides a reference for identifying organisms • Systematics or phylogeny – The study of the evolutionary history of organisms The Study of Phylogenetic Relationships • All Species Inventory (2001-2025) – To identify all species of life on Earth • Historic Background – 1735 – 1857 – 1866 – 1937 Plant and Animal Kingdoms Bacteria & fungi put in the Plant Kingdom Kingdom Protista proposed for bacteria, protozoa, algae, & fungi "Prokaryote" introduced for cells "without a nucleus" The Study of Phylogenetic Relationships – 1961 – 1959 – 1968 – 1978 – 1979 Prokaryote defined as cells in which nucleoplasm is not surrounded by a nuclear membrane Kingdom Fungi Kingdom Prokaryotae proposed Two types of prokaryotic cells found (sequence in rRNA) Three domain system proposed (Woese) above 5 kingdoms (based on the rRNA nucleotide sequence) The Three-Domain System Table 10.1 The Three-Domain System Figure 10.1 The Three-Domain System Table 10.2 Endosymbiotic Theory Figure 10.2 Figure 10.3 A Phylogenetic Hierarchy • Implies that a group of organisms evolved from a common ancestor – Some of the information used to classify comes from fossils – Fossil evidence not available for most prokaryotes – Phylogeny for prokaryotes based on DNA hybridization and rRNA sequencing Classification of Organisms • Rules for classifying and naming living organisms – several scientific entities are responsible for establishing rules • Scientific nomenclature (binomial nomenclature): genus and specific epithet (species) – Used by scientists worldwide – Bacteria or prokaryotic scientific names taken from Latin (genus can be taken from Greek) or latinized by adding suffix “-ales” & “-aceae”. – If reclassified, old name is often written in parentheses e.g. Enterococcus (Streptococcus) faecalis References •• Bergey’s Manual of Determinative •Morphology, differential Bacteriology staining, biochemical tests •Provides identification schemes for identifying bacteria and archaea •• Bergey’s Manual of Systematic Bacteriology •Provides phylogenetic information on bacteria and archaea •Based on rRNA sequencing •• Approved Lists of Bacterial •Based on published articles Names •Lists species of known prokaryotes Scientific Nomenclature Kbebsiella pneumoniae Source of Genus name Honors Edwin Klebs Source of Specific epithet The disease Pfiesteria piscicida Honors Lois Pfiester Disease in fish Scientific binomial Salmonella typhimurium Honors Daniel Salmon Stupor (typh-) in mice (muri-) Forms pus (pyo-) Streptococcus pyogenes Chains of cells (strepto-) Penicillium notatum Tuftlike (penicill-) Trypanosoma cruzi Corkscrew-like Honors Oswaldo (trypano-, borer; soma- Cruz body) Spores spread in wind (nota) Taxonomic hierarchy Figure 10.5 Species Definition • Eukaryotic species: a group of closely related organisms that breed (interbreed) among themselves but does not breed with individuals of another species • Prokaryotic species: a population of cells with similar characteristics – Clone: Population of cells derived from a single cell – Strain: Genetically different cells within a clone Domain Eukarya • Animalia: Multicellular; no cell walls; chemoheterotrophic • Plantae: Multicellular; cellulose cell walls; usually photoautotrophic • Fungi: Chemoheterotrophic; unicellular or multicellular; cell walls of chitin; develop from spores or hyphal fragments • Protista: A catchall for eukaryotic organisms that do not fit other kingdoms Prokaryotes Figure 10.6 Classification of viruses • Viral species: population of viruses with similar characteristics that occupies a particular ecological niche • Viruses are not classified as part of the three-domain system • Ecological niche of a virus is its specific host cell – Viruses may be more closely related to their hosts than other viruses Methods of Classifying and Identifying Microorganisms • Majority of Bacteria and Archaea have not been cultured – Estimate that only 1 % of these microbes have been discovered – More than 2,600 species listed in the Approved Lists of Bacterial Names, but less than 10 % are human pathogens Identification Methods • Morphological characteristics: Useful for identifying eukaryotes • Differential staining: Gram staining, acid fast staining • Biochemical tests: Determines presence of bacterial enzymes Figure 10.8 Biochemical test: numerical identification Figure 10.9 Identification Methods: Serology • Science that studies blood serum and immune responses that are evident in serum – Microorganisms are antigenic (induce/stimulate formation of antibodies when they enter an animal’s body) – Antigen (Ag): any substance that causes antibody (Ab) formation and reacts only with its specific Ab; immunogen – Antibody (Ab): proteins that circulate in the blood and combine in a highly specific way with the bacteria that caused their production Serology • Combine known antiserum + unknown bacterium • Slide agglutination • ELISA • Western blot Positive result = agglutination (clumping) Figure 10.10 Western Blot Figure 10.12 Identification Methods: Phage typing • Looks for similarities among bacteria – bacteriophages infect only specific host bacteria and usually causes lysis fo the bacterial host cells they infect • Useful in tracing the origin and course of a disease outbreak – Can trace sources of food-associated infections – Also useful for locating the source of hospital acquired (nosocomial) infection Phage Typing Figure 10.13 Identification Methods: Flow cytometry • Differences in electrical conductivity between species • Fluorescence of some species • Cells selectively stained with antibody + fluorescent dye Figure 18.11 Genetics • DNA base composition (Classification) – Comparison of the Guanine + cytosine (GC) moles % in different species can reveal the degree of species relatedness • rRNA sequencing (Classification) – Used to determine the diversity of organisms and the phylogenetic relationships among them • Polymerase Chain Reaction (PCR) – Amplification of DNA; Detect microbes that cannot be cultured by conventional methods – Used for classification & identification Identification Methods: Genetics • DNA fingerprinting – Electrophoresis of restriction enzyme digests – The more similar the patterns, the more closely related – Can be used to find sources of nosocomial infections Figure 10.14 Nucleic Acid Hybridization • Measures the ability of DNA strands from one organism to hybridize with the DNA strands of another organism – The greater the degree of hybridization, the greater the degree of relatedness (classification) – DNA-RNA hybridization can also be used – Can be used for identification (Southern blotting, DNA probe, & DNA chip) Nucleic Acid Hybridization Figure 10.15 Nucleic Acid Hybridization: DNA probe Figure 10.16 Nucleic Acid Hybridization: DNA chip Figure 10.17 Figure 10.5 Dichotomous Key • Widely used for identification purposes Chap. 10, p. 287 Cladogram • Maps that show evolutionary relationships among organisms • Made primarily using rRNA sequence Figure 10.18.1 Cladogram • Each branch point is defined by a feature shared by various species on that branch Figure 10.18.2 Chapter Review • Taxonomy: the science of classifying organisms – Provides universal names for organisms – Show degree of similarities among organisms – Provides a common reference for identifying organisms already classified • Systematics or phylogeny: the study of the evolutionary history of organisms • Provides tools for clarifying the evolution of organisms as well as their interrelationships The Three-Domain System • Proposed by Carl R. Woese in 1978 • Based on the nucleotide sequences of rRNA; a universal ancestor split into 3 lineages (endosynbiotic theory) • Eukarya: animals, fungi, plants, & protists – Eukaryotic species: a group of closely related organisms that breed (interbreed) among themselves but does not breed with individuals of another species Domain Eukarya • Animalia: Multicellular; no cell walls; chemoheterotrophic • Plantae: Multicellular; cellulose cell walls; usually photoautotrophic • Fungi: Chemoheterotrophic; unicellular or multicellular; cell walls of chitin; develop from spores or hyphal fragments • Protista: A catchall for eukaryotic organisms that do not fit other kingdoms The Three-Domain System • Bacteria: all the prokaryotes with peptidoglycan cell wall (pathogenic & nonpathogenic) • Archaea: prokaryotes that do not have peptidiglycan in their cell walls (methanogens, extreme halophiles, 7 hyperthermophiles) The Three-Domain System • Prokaryotic species: a population of cells with similar characteristics – Clone: Population of cells derived from a single cell – Strain: Genetically different cells within a clone • Phylogenetic Hierarchy – Eukaryotic evidence comes from fossils – Prokaryotic evidence comes from DNA hybridization & rRNA sequencing Viruses • Not a part of the three-domain system • Viral species: population of viruses with similar characteristics that occupies a particular ecological niche • Ecological niche of a virus is its specific host cell – Viruses may be more closely related to their hosts than other viruses Methods of Classifying and Identifying Microorganisms • Majority of Bacteria and Archaea have not been cultured – Estimate that only 1 % of these microbes have been discovered – More than 2,600 species listed in the Approved Lists of Bacterial Names, but less than 10 % are human pathogens Chapter Review • Know these terms: taxonomy, phylogeny (systematics), eukaryotic species, prokaryotic species, viral species, clone, and strain. • Know how phylogenetic hierarchy was determined (fossil evidence, rRNA sequence & DNA hybridization) Chapter Review • Know the three-domain system • Know the classifications of eukaryotes, prokaryotes, and virus. • Classification and Identification part of this chapter (some techniques are applicable to Chapter 9, too) will be tested as a take home exam (due on Friday, Oct. 17). Ten questions (matching) for the take home exam will be posted on the web and given in the class by Monday.