Chapter 19 - The inheritance of complex traits
Chapter 19
The Inheritance of Complex Traits
Quantitative Genetics
•
Former basketball star Wilt
Chamberlain (7 feet, 1 inch tall) and former renowned jockey Willie
Shoemaker (4 feet, 11 inches tall) show some of the extremes in human height—a quantitative trait
•
Quantitative traits
•
Statistics
•
Phenotypic distributions
•
Reaction norms
•
Broad-sense heritability
•
Narrow-sense heritability
•
QTL mapping
What is quantitative genetics?
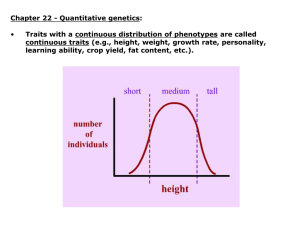
• Traits such as height that show a continuous range of variation and do not behave in a simple Mendelian fashion are known as quantitiative or complex traits
• Calculation of a mean (average) value and variance/standard deviation
• Quantitative trait loci (QTLs) produce continuously variable phenotypes
Basic statistical concepts
• phenotypic variation in quantitative traits described statistically (frequency histogram)
• features of statistical distributions:
– central tendency (mean or average)—observed values around a particular point
– dispersion (variance or standard deviation)— how much variation about the mean
Populations can be described mathematically
Mean of a population X= 1/n ΣX
Populations can be described mathematically
-Variance of a population
V x
= 1/n Σ (X –X) 2
The standard deviation is the square root of the variance
In a normal distribution, the standard deviation describes the distribution.
How can the relative contributions to Yao Ming ’ s height be determined?
If we could clone Yao Ming and raise his clones in different environments, we could determine the relative contributions of genetics and environment.
What is quantitative genetics?
• Correlates phenotypic trait distributions with genotypes, environment
• Some questions researchers ask:
– What proportion of observed phenotypic variation is determined by genetic variation?
– What proportion of observed phenotypic variation is determined by environmental variation?
– Do different alleles for a gene produce different effects?
– What phenotypes of the genotypes inhabit different environments?
– How many loci are involved for a trait?
– What offspring result from crosses of different quantitative phenotypes?
Quantitative traits described by frequency distribution several genes?
few genes?
# genes partly determines curve “ smoothness ” many genes?
Reaction norm relates environment and phenotype
Reaction norm relates environment and phenotype
• reaction norm = relation between environment and phenotype for a particular genotype
• can graph this
• under a “ distribution of environments ” , any given genotype yields a “ distribution of phenotypes ”
Reaction norm shows no genotype
“ best ” for all environments
Reaction norm shows no genotype
“ best ” for all environments
• few reaction norm studies to date on quantitative traits of wild species
• many on domesticated crops (e.g., corn, strawberries)
• no genotype consistently produces “ superior ” phenotypes across all environmental conditions
Broad-Sense Heritability:
Nature Versus Nurture
• If trait is heritable, we can quantify heritability
• We can separate total phenotypic variation of population ( V
X
) into genetic variance ( V g
) and environmental variance ( V e
)
• Broad-sense heritability (
H 2 ) is defined as H 2 = V g
/V
X
•
H 2 varies from 0 (all environment) to 1 (all genetic)
Monozygotic twins are genetically identical
Narrow–Sense Heritability
• understanding of broad-sense heritability ( H 2 ) useful, but may want to know more about genetic variance specifically
• heterozygotes not exactly intermediate in phenotype between homozygotes (partial dominance)
• difference in average effect between alleles is
“ additive effect ”
• accounts for some, but not all, variance in phenotype
Narrow-sense heritability
• so, genetic variance (
V g
) can be subdivided into additive genetic variation ( V a
) and dominance variance ( V d
): V g
= V a
+ V d
• recall that total phenotypic variance (
V x
) is
V x
= V g
+ V e
= V a
+ V d
+ V e
• so, narrow-sense heritability ( h 2 ) is defined as: h 2 = V a
/ V x
The difference between additive and dominant gene action
Estimating components of genetic variance
• allows us to use h 2 to predict effects of artificial selection
• Animal and plant breeders use h 2
Different populations have different heritabilities for traits
QTL Mapping
Mapping quantitative loci requires:
1.
Setting up a cross that will result in a segregating population
2.
Developing assays for a large number of molecular markers in the organism
3.
Assaying for correlation between the trait in question and the molecular markers in offspring
Association mapping finds a gene for body size in dogs
Summary
• reaction norm studies show that no single genotype
“ superior ” over all environments
• broad heritability (
H 2 ) separates genotypic from environmentally induced variance: H 2 = V g
/ V x
• narrow heritability ( h 2 ) subdivides genetic variance into additive and dominance variance: h 2 = V a
/ V x
• can use h 2 to predict effects of artificial selection
• can use various two mapping approaches to determine the genetic basis of quantitative traits
• many traits have many contributing loci, each usually providing small effects