Molecular Modeling of Crystal Structures
advertisement

Molecular Modeling
of Crystal Structures
molecules
surfaces
crystals
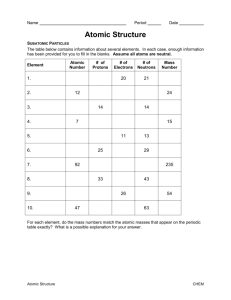
1. Potential energy functions
QM ab initio: distribution of electrons over the system.
Gaussian94, Gamess, ...
Semi-empirical methods: pre-calculated values or neglect
of some parts of the ab-initio calculation.
MOPAC (mopac6, 7, 93, 2000)
Empirical methods: observed/fitted values for interactions
between atoms.
Sybyl, Cerius2, Gromos, ...
Potential energy functions
Differences:
* Speed (as a function of system size)
* Accuracy
* Intended use (heat of fusion; conformational energies;
transition states; vibrations/spectra; …)
* Transferability / applicability
* Availability / user interface
Potential energy functions
Focus: Molecular Mechanics (MM)
“Ball and Spring” model of molecules, based on simple equations
giving U as function of atomic coordinates
G = U + pV - TS
H = U + pV
EMM = U
Molecular Mechanics
system from atoms + bonds
EMM = Estretch + Ebend + Etorsion + Evdw + Ecoul + ...
bonded
• stretching
• bending
• torsion
H
non-bonded
H
H
C
C
H
H
H
MM: interactions via bonds
r
bond stretch
Es = 1/2 ks(r-r0)2
… + C3(r-r0)3 + C4(r-r0)4
E
- True
.. modeled via (r-r0)2
r0
r
Force field parameters: bond lengths (Dreiding)
C
Es = 1/2 ks(r-r0)2
C
Bond type
r0 (Å) ks (kcal/mol.A2)
C(sp3)--C(sp3)
C(sp3)--C(sp2)
C(sp2)--C(sp2)
C(sp3)--H
1.53 700
1.43 700
1.33 1400
1.09 700
C
C
E
kS=700: E=3 kcal ~ r=0.09Å
MM: interactions via bonds
bending
Eb = 1/2 kb(-0)2
E
0
Force field parameters: bond angles (Dreiding)
C
O
H
E
Angle type
0 (°)
X--C(sp3)--X
X--O(sp3)--X
109.471 100
104.510 100
kb(kcal/mol.rad2)
E=3 kcal ~ =14°
Force field parameters: torsion angles (Dreiding)
Etor = V1[1 - cos (-01) ]
V2[1 - cos 2(-02)]
V3[1 - cos 3(-03)]
C
E
V3
0
60
120
180
C
Force field parameters: torsion angles (Dreiding)
C
C
torsion type
C
C
n V (kcal/mol) 0 (°)
X--C(sp3)--C(sp3)--X 3 1.0
X--C(sp2)--C(sp2)--X 2 22.5
180
0
Non-bonded interactions: Van der Waals
repulsive: ~r-10
attractive: ~r-6
E=D0[(r0/r)12-2(r0/r)6]
(Lennard-Jones)
E=D0{exp[a(r0/r)]-b(r0/r)6}
(Buckingham; “exp-6”)
Non-bonded interactions: Coulomb
(electrostatic)
atomic partial charges:
+
+
-
Eij=(qixqj)/(rij)
+
+
atomic/molecular multipoles:
E=ixj/Dr3
additional energy terms in force fields
* out-of-plane energy term
* Hydrogen bond energy term
MM energy calculation
EMM = Estretch + Ebend + Etorsion + Evdw + Ecoul + ...
bonded
1
non-bonded
5
2
3
4
6
bonded non-bonded
1…2
1…3
1…4
1…4: scaled
1…5
1…6/7/8
8
7
Some available force fields
FF
software
Gromos Gromos
Charmm Charmm; Quanta
Amber Amber
Tripos
Sybyl
Dreiding Cerius
Compass Cerius
CVFF
Cerius
Glass2.01 Cerius
focus
bio
bio
bio
general
general
general
general
ionic
Force field parameters:
where do they come from?
1. Mimic physical properties of individual elements or atom types,
producing a “physical” force field.
Properties can be taken from experimental data, or ab-initio
calculations.
Examples: Dreiding, Compass.
+ outcome will be ‘reasonable’, predictable; extension to new
systems relatively straightforward.
- performance not very good.
Force field parameters:
where do they come from?
2. Optimize all parameters with respect to a set of test data,
producing a “consistent” force field.
Test set can be chosen to represent the system under investigation.
Examples: CFF, CVFF.
+ outcome often good for a particular type of systems, or a
particular property (e.g. IR spectrum).
- extension to new systems can be difficult; no direct link to
‘physical reality’
Force field parameters:
where do they come from?
3. Apply common sense and look at what the neighbors do.
Examples: Gromos.
+ does not waste time on FF parameterization;
resonable results.
-?
Atomic charges
Why?
To include the effect of the charge distribution over the system.
Some sp2 oxygens are
more negative than others.
How?
Assign a small charge to each atom.
Caveat: interaction with other force field parameters (e.g. VdW).
Atomic charges
What is the atomic charge?
* Based on atomic electronegativity, optimized for a given FF.
example: Gasteiger charges.
•Based on atomic electronegativity and the resulting electrical field.
example: Charge Equilibrium charges (QEq).
* Based on the electronic distribution calculated by QM.
example: Mulliken charges.
* Based on the electrostatic potential near the molecule,
calculated by a non-empirical method (or determined experimentally).
examples: Chelp, ChelpG, RESP.
Atomic charges
Properties and features of different charge schemes:
* Depends on molecular conformation?
* Easy (=quick) to calculate?
* Performance in combination with force field?
Known-to-be-good combinations:
Tripos -- Gasteiger
Dreiding -- ESP
Compass -- Compass
Atomic charges:
charges fitted to the ElectroStatic Potential (ESP)
mechanism:
Coulomb interactions result from the electrostatic potential
around a molecule.
+
+ +
+
+ H - -- O
-- + H +
+
+
+
H+
Atomic charges:
charges fitted to the ElectroStatic Potential (ESP)
sample point
molecule
H
QM
wave function
electron density
O
H
sample true ESP
mathematical fit
atomic charges that
reproduce the true ESP
for each sample point:
atomsq/r= ESPQM
* atomic q as variables
Atomic charges:
charges fitted to the ElectroStatic Potential (ESP)
Properties and features of different fitting schemes:
* Number of sample points.
* Position of sample points.
* Additional restraints (e.g. all qH in CH3 equal).
* Fitting to multiple conformations.
Known-to-be-good fitting schemes:
ChelpG
RESP