The Mechanism of Translation II: Elongation and Termination
advertisement
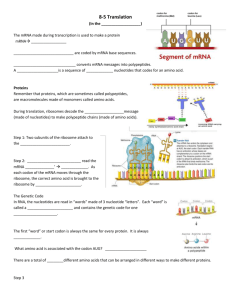
The Mechanism of Translation II: Elongation and Termination Chapter 18 Direction of Polypeptide Synthesis and mRNA Translation • Messenger RNAs are read in the 5’3’ direction • Proteins are made in the aminocarboxyl direction - means that the amino terminal amino acid is added first The Genetic Code • The term genetic code refers to the set of 3base code words (codons) in mRNA that represent the 20 amino acids in proteins • Nonoverlapping codons - Each base is part of at most one codon in nonoverlapping codons - In an overlapping code - one base may be part of two or even three codons The Triplet Code • The genetic code is a set of three-base code words - or codons – In mRNA - codons instruct the ribosome to incorporate specific amino acids into a polypeptide • Code is nonoverlapping – Each base is part of only one codon • Devoid of gaps or commas – Each base in the coding region of an mRNA is part of a codon Coding Properties of Synthetic mRNAs The Genetic Code • There are 64 codons – 3 are stop signals – Remainder code for amino acids – The genetic code is highly degenerate Unusual Base Pairs Between Codon and Anticodon Degeneracy of genetic code is accommodated by: – Isoaccepting species of tRNA: bind same amino acid but recognize different codons – Wobble - the 3rd base of a codon is allowed to move slightly from its normal position to form a nonWatson-Crick base pair with the anticodon – Wobble allows same aminoacyl-tRNA to pair with more than one codon Compare standard Watson-Crick base pairing with wobble base pairs • Wobble pairs are: – G-U – I-A Deviations from “Universal” Genetic Code The (Almost) universal code • Genetic code is NOT strictly universal • Transitions and Transversions • Certain eukaryotic nuclei and mitochondria along with at least one bacterium – Codons cause termination in standard genetic code can code for amino acids Trp, Glu – Mitochondrial genomes and nuclei of at least one yeast have sense of codon changed from one amino acid to another Elongation mechanism in E.coli Elongation takes place in three steps: 1. EF-Tu with GTP binds aminoacyl-tRNA to the ribosomal A site (empty – based on second codon). 2. Peptidyl transferase forms a peptide bond between peptide in P site and newly arrived aminoacyl-tRNA in the A site Lengthens peptide by one amino acid and shifts it to the A site Elongation mechanism in E.coli Translocation • EF-G with GTP translocates the growing peptidyl-tRNA with its mRNA codon to the P site - The deacylated tRNA in P site leaves ribosome via E site. - The dipeptidyl-tRNA in A site along with its corresponding codon moves into P site - Steps keep on repeating Protein Factors and Peptide Bond Formation • One factor is T- transfer – It transfers aminoacyl-tRNAs to the ribosome – has 2 different proteins • Tu - u stands for unstable • Ts - s stands for stable • Second factor is G - GTPase activity • Factors EF-Tu and EF-Ts are involved in the first elongation step • Factor EF-g participates in the third step Elongation Step 2 • One the initiation factors and EF-Tu have done their jobs - the ribosome has fMet-tRNA in the P site and aminoacyl-tRNA in the A site • Now form the first peptide bond • No new elongation factors participate in this event • Ribosome contains the enzymatic activity peptidyl transferase - that forms peptide bond Peptide Bond Formation • The peptidyl transferase resides on the 50S ribosomal particle • Minimum components necessary for activity are 23S rRNA and proteins L2 and L3 • 23S rRNA is at the catalytic center of peptidyl transferase Elongation Step 3 • When peptidyl transferase has worked: – Ribosome has peptidyl-tRNA in the A site – Deacylated tRNA in the P site • Translocation - moves mRNA and peptidyltRNA one codon’s length through the ribosome – Places peptidyl-tRNA in the P site – Ejects the deacylated tRNA – Process requires elongation factor EF-G which hydrolyzes GTP after translocation is complete Proofreading • Protein synthesis accuracy comes from charging tRNAs with correct amino acids • Proofreading is correcting translation by rejecting an incorrect aminoacyl-tRNA before it can donate its amino acid • Protein-synthesizing machinery achieves accuracy during elongation in two steps Protein-Synthesizing Machinery • Two steps achieve accuracy: – Gets rid of ternary complexes bearing wrong aminoacyltRNA before GTP hydrolysis – If this screen fails, still eliminate incorrect aminoacyltRNA in the proofreading step before wrong amino acid is incorporated into growing protein chain • Steps rely on weakness of incorrect codon-anticodon base pairing to ensure dissociation occurs more rapidly than either GTP hydrolysis or peptide bond formation Proofreading Balance • Balance between speed and accuracy of translation is delicate – If peptide bond formation goes too fast • Incorrect aminoacyl-tRNAs do not have enough time to leave the ribosome • Incorrect amino acids are incorporated into proteins – If translation goes too slowly • Proteins are not made fast enough for the organism to grow successfully • Actual error rate, ~0.01% per amino acid is a good balance between speed and accuracy Three-Nucleotide Movement Each translocation event moves the mRNA on codon length, or 3 nt through the ribosome Role of GTP and EF-G • GTP and EF-G are necessary for translocation – Translocation activity appears to be inherent in the ribosome – This activity can be expressed without EF-G and GTP • GTP hydrolysis – Precedes translocation – Significantly accelerate translocation • New round of elongation occurs if: – EF-G must be released from the ribosome – Release depends on GTP hydrolysis Termination • Elongation cycle repeats over and over – Adds amino acids one at a time – Grows the polypeptide product • Finally ribosome encounters a stop codon UAG, UAA, UGA – Stop codon signals time for last step – Translation last step is termination Termination Mutations • Mutations can create termination codons within an mRNA causing premature termination of translation – Amber mutation creates UAG – Ochre mutation creates UAA – Opal mutation creates UGA Termination Mutations • Amber mutations are caused by mutagens that give rise to missense mutations • Ochre and opal mutations do not respond to the same suppressors as do the amber mutations – Ochre mutations have their own suppressors – Opal mutations also have unique suppressors Release Factors • Prokaryotic translation termination is mediated by 3 factors: – RF1 recognizes UAA and UAG – RF2 recognizes UAA and UGA – RF3 is a GTP-binding protein facilitating binding of RF1 and RF2 to the ribosome • Eukaryotes has 2 release factors: – eRF1 recognizes all 3 termination codons – eRF3 is a ribosome-dependent GTPase helping eRF1 release the finished polypeptide Dealing with Aberrant Termination • Two kinds of aberrant mRNAs can lead to aberrant termination – Nonsense mutations can occur that cause premature termination – Some mRNAs (non-stop mRNAs) lack termination codons • Synthesis of mRNA was aborted upstream of termination codon • Ribosomes translate through non-stop mRNAs and then stall • Both events cause problems in the cell yielding incomplete proteins with adverse effects on the cell Non-Stop mRNAs • Prokaryotes deal with non-stop mRNAs by tmRNA-mediated ribosome rescue – tmRNA are about 300 nt long – 5’- and 3’-ends come together to form a tRNAlike domain (TLD) resembling a tRNA Non-Stop mRNAs • Prokaryotes deal with non-stop mRNAs by tmRNAmediated ribosome rescue – Alanyl-tmRNA resembles alanyl-tRNA – Binds to vacant A site of a ribosome stalled on a non-stop mRNA – Donates its alanine to the stalled polypeptide • Ribosome shifts to translating an ORF on the tmRNA (transfer-messenger RNA) – Adds another 9 amino acids to the polypeptide before terminating – Extra amino acids target the polypeptide for destruction – Nuclease destroys non-stop mRNA Eukaryotic Aberrant Termination • Eukaryotes do not have tmRNA • Eukaryotic ribosomes stalled at the end of the poly(A) tail contain 0 – 3 nt of poly(A) tail – This stalled ribosome state is recognized by carboxyl-terminal domain of a protein called Ski7p – Ski7p also associates tightly with cytoplasmic exosome, cousin of nuclear exosome – Non-stop mRNA recruit Ski7p-exosome complex to the vacant A site – Ski complex is recruited to the A site Exosome-Mediated Degradation • Exosome, positioned just at the end of non-stop mRNA, degrades that RNA • Aberrant polypeptide is presumably destroyed Posttranslation • Translation events do not end with termination – Proteins must fold properly – Ribosomes need to be released from mRNA and engage in further translation rounds • Folding is actually a cotranslational event occurring as nascent polypeptide is being made Folding Nascent Proteins • Most newly-made polypeptides do not fold properly alone – Polypeptides require folding help from molecular chaperones – E. coli cells use a trigger factor • Associates with the large ribosomal subunit • Catches the nascent polypeptide emerging from ribosomal exit tunnel in a hydrophobic basket to protect from water – Archaea and eukaryotes lack trigger factor- use freestanding chaperones Release of Ribosomes from mRNA • Help is required from ribosome recycling factor (RRF) and EF-G – RRF resembles a tRNA • Binds to ribosome A site • Uses a position not normally taken by a tRNA – Collaborates with EF-G in releasing either 50S ribosome subunit or whole ribosome • • • This project is funded by a grant awarded under the President’s Community Based Job Training Grant as implemented by the U.S. Department of Labor’s Employment and Training Administration (CB-15-162-06-60). NCC is an equal opportunity employer and does not discriminate on the following basis: against any individual in the United States, on the basis of race, color, religion, sex, national origin, age disability, political affiliation or belief; and against any beneficiary of programs financially assisted under Title I of the Workforce Investment Act of 1998 (WIA), on the basis of the beneficiary’s citizenship/status as a lawfully admitted immigrant authorized to work in the United States, or his or her participation in any WIA Title I-financially assisted program or activity. Disclaimer • This workforce solution was funded by a grant awarded under the President’s Community-Based Job Training Grants as implemented by the U.S. Department of Labor’s Employment and Training Administration. The solution was created by the grantee and does not necessarily reflect the official position of the U.S. Department of Labor. The Department of Labor makes no guarantees, warranties, or assurances of any kind, express or implied, with respect to such information, including any information on linked sites and including, but not limited to, accuracy of the information or its completeness, timeliness, usefulness, adequacy, continued availability, or ownership. This solution is copyrighted by the institution that created it. Internal use by an organization and/or personal use by an individual for non-commercial purposes is permissible. All other uses require the prior authorization of the copyright owner.