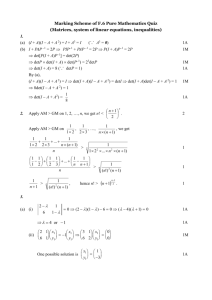
Text Mining Overview
advertisement

Text mining in the field of evolutionary biology: facilitating scholarly collaboration Sarah Carrier February 2008 What is text mining? • Deriving novel, relevant information from unstructured information (text). • Identification of patterns and trends. • Typical techniques: – Clustering – Categorization – Concept/entity extraction -> dictionary-based, statistical methods/machine learning – Document summarization Long-Term Objective 1. To identify biological entities through text mining methods, then categorize them into predetermined classes of objects 2. To describe biological concepts using simple ontologies - for example, use the controlled vocabulary generated in step 1 to describe results and methods Semester Objective 1. To categorize evolutionary biology abstracts into 5 different predetermined categories using nouns and noun-phrases associated with the text. 2. To prepare for long-term objectives. Motivation • Scholarly collaboration • Generation of ontologies to describe results of experiments, to enhance meta-analyses for research purposes • Web publishing • Indexing by central repositories Motivation and Current Research • need in the life sciences for alternatives to keywordbased approaches based in the traditional information retrieval framework • extensive (text mining) work is being done to identify protein-protein interactions and gene annotations • extracted entities can be linked to existing ontologies and potentially used to generate new ontologies • the most common text mining applications in the life sciences tend toward information extraction, as this method produces a potential solution to the deluge of information in the field Manual Keyword Identification • 8 categories: concept, field/discipline, gene, habitat, method, place, taxon, time period • 104 articles, 5 journals, 600 keywords - 551 with duplicates removed, most terms ended up in the “concept” category -> varied sizes • Manual categorization accomplished with domain experts on the Dryad team, matched with existing terminologies • 16% were duplicates, avg. 50% matched terminologies - implies that controlled vocabularies should be used for standardization Some potential challenges • Evolutionary biology is an interdisciplinary field: ecology, genomics, paleontology, population genetics, physiology, systematics • A varied and complex terminology for the life sciences • Incredibly sparse dataset • Coverage of existing terminologies incomplete (UMLS, Open Biomedical Ontologies) Methodology • MEDLINE abstracts from American Naturalist, Ecology, Journal of Evolutionary Biology, Molecular Ecology, Molecular Biology and Evolution, Systematic Biology • Total: 15,179 abstracts, 227,731 terms extracted from list of MeSH terms and 831,245 terms using abstract • Standard preprocessing of abstracts using Perl, including the Porter stemmer and the Brill Tagger An Example PMID- 17206577 TI- Ecological specialization and adaptive decay in digital organisms. AB- The transition from generalist to specialist may entail the loss of unused traits or abilities, resulting in narrow niche breadth. Here we examine the process of specialization in digital organisms--selfreplicating computer programs that mutate, adapt, and evolve. Digital organisms obtain energy by performing computations with numbers they input from their environment. We examined the evolutionary trajectory of generalist organisms in an ecologically narrow environment, where only a single computation yielded energy. CONTINUED… MH- *Adaptation, Biological, Competitive Behavior, Computer Simulation, Ecology, *Evolution, Molecular, Genotype, *Models, Genetic, Mutation, Phenotype, Software Preprocessing 17206577|1|transition 17206577|1|specialist 17206577|1|loss of unus trait 17206577|1|trait 17206577|1|generalist 17206577|1|loss 17206577|1|transition from generalist 17206577|1|unus trait 17206577|1|narrow nich breadth 17206577|1|nich breadth 17206577|1|breadth 17206577|2|process 17206577|2|abil 17206577|2|nich • CONCEPT: regressive evolution, specialization, pleiotropy, adaptation, mutation accumulation • METHOD: digital evolution Preprocessing, cont. The/DET transition/NN from/IN generalist/NN to/TO specialist/NN may/MD entail/VB the/DET loss/NN of/IN unused/JJ traits/NNS or/CC abilities/NNS ,/PPC resulting/VBG in/IN narrow/JJ niche/NN breadth/NN ./PP Here/RB we/PRP examine/VBP the/DET process/NN of/IN specialization/NN in/IN digital/JJ organisms/NNS selfreplicating/NN computer/NN programs/NNS that/IN mutate/VB ,/PPC adapt/VBP ,/PPC and/CC evolve/VB ./PP Digital/NNP organisms/NNS obtain/VBP energy/NN by/IN performing/VBG computations/NNS with/IN numbers/NNS they/PRP input/NN from/IN their/PRPS environment/NN ./PP We/PRP examined/VBD the/DET evolutionary/JJ trajectory/NN of/IN generalist/NN organisms/NNS in/IN an/DET ecologically/RB narrow/JJ environment/NN ,/PPC where/WRB only/RB a/DET single/JJ computation/NN yielded/VBD energy/NN ./PP We/PRP determined/VBD the/DET extent/NN to/TO which/WDT An Example • <MeshHeadingList> • <MeshHeading> • <DescriptorName MajorTopicYN="N">Adaptation, Physiological • </DescriptorName> • </MeshHeading> • <QualifierName MajorTopicYN="N">genetics • </QualifierName> • <QualifierName MajorTopicYN="Y">metabolism • </QualifierName> • </MeshHeading> • <MeshHeading> • <DescriptorName MajorTopicYN="N">Predatory Behavior • </DescriptorName> • </MeshHeading> • </MeshHeadingList> Most Frequent Abstrac t Terms (collection) speci popul gene result sequenc studi data ana lysi pattern evo lut variat dna phylogene t region model leve l rate ana lys structur select Most Frequent MeSH Terms (collection) gene t sequenc anim dna phys iologi phylogen i evo lut popul data ana lysi model molecular sequenc da ta sequenc da ta gene variat acid base base sequenc classif protein Other Steps • TF*IDF weighting, pruning – Challenges: skew in category sizes (“concept” being the largest), lack of truly discriminative terms • Application of a machine-learning model: Hidden Markov Models, Support Vector Machines – SVMs: outperform HMM • also better for large, sparse datasets • Evaluation: – Recall, Precision, F-Scores – Presentation to Dryad domain experts for feedback Future Steps • Use of existing vocabularies to assist in controlling terminology: NBII thesaurus, MeSH, GTN, WordNet, Gene Ontology, ITIS, UBIO, UMLS, etc. Ontology generation? • The POS processing has already been done - the verb is an essential element of the relationship • Find most common verbs and define them as “relational verbs” • Methodology: using POS tags, pull out “triplets” or certain sequences of words – NOUN - VERB - NOUN …in some studies, prepositions are also analyzed Ontology, cont. Our/PRPS results/NNS show/VBP that/IN as/IN organisms/NNS evolved/VBD improved/VBN performance/NN of/IN the/DET selected/JJ function/NN ,/PPC they/PRP often/RB lost/VBN the/DET ability/NN to/TO perform/VB other/JJ computations/NNS ,/PPC and/CC these/DET losses/NNS resulted/VBD most/JJS often/RB from/IN the/DET accumulation/NN of/IN neutral/JJ and/CC deleterious/JJ mutations/NNS ./PP Conclusions • Term variation and ambiguity presented a challenge in my project because it yielded a very sparse data set • With more time I would have supplemented the dataset I generated this semester with more data from more abstracts, perhaps even the full text, if available • Although the objective of the project changed over the semester, the results provide valuable insight into the structure and use of evolutionary biology vocabularies • Potential future developments in the project, namely ontology generation, would have a positive impact on scholarly communication amongst researchers in the field of evolutionary biology Thank you!