Lab meeting on GWUE, Feb 2005
advertisement
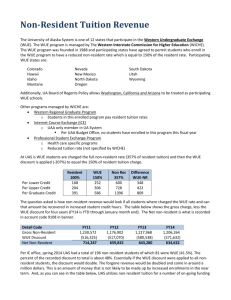
Genomic analysis of water use efficiency Boyce Thompson Institute for Plant Science Cornell University Oklahoma State University University of North Carolina at Chapel Hill http://isotope.bti.cornell.edu/ Collaborators • Cornell/Boyce Thompson: Jonathan Comstock, Susan McCouch – – – – – Christine Fleet Roman Pausch Wendy Vonhof Shiqin Xu Yunbi Xu • Oklahoma State: Bjorn Martin, Chuck Tauer – Shakuntala Fathepure – Baige Zhao • UNC Chapel Hill: Todd Vision – Maria Tsompana – Lindsey Swanson Water use efficiency • A fundamental trade-off for plants – Open stomates allow photosynthesis – But also result in water loss • WUE is the ratio of carbon fixed to water lost – Somewhat related to drought tolerance – More closely to yield potential under irrigation • Water is the most limiting resource to global agricultural production • In some crops, and under some conditions, greater WUE would be desirable and in others less Three levels of WUE • Whole-field (under agronomic control) • Whole-plant (driven by respiration) • Single-leaf (focus here) Leaf-level WUE sun wind ci wi ca H2O wa CO2 photosynthesis ca ci WUE transpiration 1.6wa w i The challenges of working with WUE • WUE is a complex trait – Rarely if ever controlled by a single gene – Very sensitive to environment • Breeding for WUE has not worked – Too many deleterious side-effects • We know almost nothing about the molecular biology of how plants adjust their WUE – Could we engineer WUE if we knew more? • QTL mapping as a “foot in the door” to discover the pathways involved in WUE Quantitative trait loci (QTL) P1 (+) P2 (-) F1 (0) F2 + + 0 LOD Stable carbon isotopes • Direct physiological measurement of WUE is not quick and cheap enough for QTL studies a proxy is needed • Stable isotopes are naturally occuring – Atmospheric CO2 is 99 12C : 1 13C • Rubisco, the key enzyme in carbon fixation, discriminates against 13C • Easily measured by mass spectrometry Isotope measurements • Isotopic ratio R = 13C/12C • Discrimination index D = (Rair/Rplant) – 1 D and WUE • Both ∆ & WUE depend on the CO2 diffusion gradient • In C3 plants, variation in this gradient is the primary determinant of D and leaf-level WUE. • D provides a high-throughput proxy for ci – Values of D are typically negative – Values closer to zero represent greater WUE (more carbon fixed per unit of water) Goals • To dissect natural variation in WUE • Discovery and characterization of WUE quantitative trait loci (QTL) – Rice (upland vs rice paddy cultivation) – Tomato (desert versus cultivated species) • Lay ground-work for positional cloning – Fine mapping – Introgression lines Survey of variability in rice • Assayed variation in D among – Landraces and elite cultivars – Related wild species – The offspring of four wide crosses • • • • Lamont x Teqing Kasalath x Nipponbare IR64 x Nipponbare O. rufipogon x Jefferson • Variation in the offspring of a single cross can be as wide as the variation among all cultivated/wild accessions! • Upland/lowland distinction not that helpful… Survey of variability in rice LOD=8.60 WUE QTL On Chromosome 1 Genetic Map Genomic sequence www.gramene.org Mapping WUE QTL in tomato • Wild desert species of tomato (e.g. Solanum pennellii) have high WUE relative to cultivated species (S. lycopersicon) • On the minus side – The genome sequence is not available yet • On the plus side – Zamir introgression lines for S. lycopersicon x S. pennellii greatly facilitate mapping QTL in pennellii population Possible physiological basis for WUE • Several of the candidate QTL lines have – High nitrogen content = abundant protein – Low specific leaf area (m2/g) • These correlates suggest that increased carboxylation capacity may be responsible for greater WUE in these QTL Finding crossovers within IL5-4 • QTL can be located more precisely if IL5-4 introgression can be broken up • Backcrossed IL5-4 to cultivated parent • Genotyped F2 progeny for flanking markers QuickTime™ and a TIFF (LZW) decompressor are needed to see this picture. Principle of fine-mapping (Mendelization) mm mm qq mm mm mm mm mm qq mm mm flanking marker 1 QTL qq internal marker 1 flanking marker 2 Fine-mapping IL5-4 QTL • 16 crossovers obtained from ~2000 backcross F2 plants • These were selfed to produce backcross F3s – D values obtained for F3 plants • Scoring internal STS markers – These allow us to align to the tomato physical map – One internal STS marker done – Several more in development • AFLP markers are currently being mapped – Not physically mapped, but abundant and easy to score IL5-3 TG351 72.7 TG351 76.2 TG60, CT80 75 TG60, CT80 78.4 CP58B, CHS3 77.2 CP58B, CHS3 IL5-4 QTL 73.9 104 TG60 105 T1777 106 86.1 CD78 84.9 CD78 88.7 TG69 87.5 TG69 IL Population F2 1992 SSR590, T1541 108 T1584 111 TG69 F2 2000 PCR length polymorphism already scored SSR marker available dCAPS marker available Screening for polymorphisms (1 or more introns predicted) Screening for polymorphisms (no intron predicted) Primers under development TG69 physical contig Now what? • Adding additional STS to IL5-4 (UNC) – Goal is <1cM (=1 Mb) resolution • Identifying BAC contigs containing markers in QTL candidate region (UNC) – BAC skimming to obtain high density markers – Comparative mapping in Arabidopsis for candidate gene analysis • Generating overlapping congenic lines in IL5-4 by marker assisted selection (OSU)