7.3: Translation
advertisement

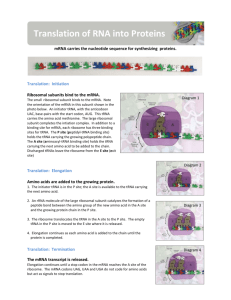
7.3: Translation Ribosome review… What are ribosomes composed of? How many parts are the to a ribosome during translation? How many tRNA binding sites are inside the ribosome? What are they called? tRNA structure Referred to as the ‘clover leaf’ structure (fig. 3, p.g. 363) On your diagram, highlight the anticodon and the site for attaching an amino acid. What bonds exist inside the tRNA molecule? What is an advantage of having this type of bond in this location? Enzymes A enzyme is a biological catalyst. *remember* enzymes are highly specific, each enzyme will only function at a optimum temperature, pH and each will bond with a specific molecule = substrate. tRNA activating enzymes There are 20 different tRNA activating enzymes. Why? Fig. 5, pg. 365. shows a tRNA molecule ‘collecting’ an amino acid via the tRNA activating enzyme. Energy is required for this process. Energy is supplied by ATP attaching to the enzyme at the same time as the amino acid. The ATP is then hydrolysed, which will provide the necessary energy for the formation of a covalent bond between the amino acid and the incoming tRNA. Once the bonding is complete, the activated tRNA exits the enzyme along with an AMP molecule. Stages of translation Initiation Elongation (some times referred to as elongation/translocation phase) Termination Use the diagrams/notes on pg. 364/5 to define these stages. 1. Initiation - The process starting An activated amino acid (methionine) attached to a tRNA with the anitcodon UAC. Combines with a small ribosomal subunit and an mRNA strand. A large ribosomal subunit combines as well, producing a translation initiation complex. Initiation factors are proteins which join the complex together. Subunits moves down mRNA strand until start codon is reached (AUG). Hydrogen bonds form between the initiator tRNA and the start codon. Energy in this case does not come from ATP. It comes from Guanosine triphosphate (GTP) which is another energy rich molecule – very similar to ATP. 2. Elongation - the chain growing tRNA’s bring amino acids to the mRNA-ribosomal complex. Order specified by the codons. Proteins called elongation factors assists binding tRNA’s to the mRNA codons at site A. Initiator tRNA then moves to site P. Ribosomes catalyse the formation of peptide bonds between amino acids. 3. Translocation tRNA relocating within the ribosome Takes place during elongation tRNA moving from site A P E. Occurs in a 5’ to 3’ direction so ribosomal complex is moving along the mRNA strand towards the 3’ end. 4. Termination The process ending There are three stop codons. When one reaches site A, a release factor (protein) fills site A. The release factor does not carry amino acid. It causes the bond linking the the tRNA to the P site to hydrolyse. Releasing the polypeptide from the ribosome. The ribosome then separates form the mRNA and splits into it’s subunits. More about ribosomes… Described as either free or bound. FREE = produce proteins for use within the cell (cytoplasm, mitochondria, chloroplast) translation takes place in the cytoplasm (cytosol) or the endoplasmic reticulum. BOUND = produce proteins which will be secreted or used in the lysosomes. Translation occurs on ribosomes attached to the ER. Signal sequences on the amino acid being translated determines the site of translation. See fig. 11, pg. 366. Polysomes When more than one ribosome is attached to a single mRNA. In prokaryotes, polysomes appear immediately after transcription. Why? In eukaryotes, polysomes appear in either the cytoplasm or next to the ER Protein structure Primary organization A simple chain of amino acids attached by peptide bonds. Polypeptide chains can have hundreds of amino acids. Primary structure influences the next three structures. Changing just one amino acids can have drastic effects of the structure of proteins. Sickle cell anaemia - when a single amino acid in haemoglobin is changed. Result - RBC no longer able to carry oxygen. Secondary organization Hydrogen bonds form between oxygen from the carboxyl group on one amino acid and the hydrogen from the amino group on another amino acid. Most common secondary structure are the alphahelix and beta-pleated sheets. Regular repeating pattern. Sketch fig. 16 pg. 370. The Bonds Bond Description Covalent Between sulphur atoms, forming disulphide bridges – very strong Between polar side chains Hydrogen Van der Waals Interactions among hydrophobic side chains of amino acids. Strong interactions (hydrophobic side chains forced inwards, when hydrophilic chains interact with water on the outside of the molecule) Ionic Between positive and negative side chains Tertiary organization Polypeptide folds and coils to form a complex molecular shape (3D) Caused by interactions between R groups; including H-bonds,Van der Waals, disulphide bridges, ionic bonds and hydrophilic / hydrophobic interactions Tertiary structure may be important for the function of the enzyme (e.g. specificity of active site in enzymes) Quartenary organization Interactions between multiple polypeptides or prosthetic groups that results in a single, larger, biologically active protein A prosthetic group is an inorganic compound involved in protein structure or function (e.g. the heme group in haemoglobin) A protein containing a prosthetic group is called a conjugated protein Quaternary structure may be held together by a variety of bonds (similar to tertiary structure) Protein Data Bank (PDB) Google protein data bank, or use this link http://www.rcsb.org/pdb/home/home.do Search for either thermus thermophilus ribosome or tRNA ribosome. See if you can view the large and small subunits using this software. Also available on the website (companion to the textbook!) www.oxfordsecondary.co.uk/ib-biology