Nimrod-G - Rajkumar Buyya
advertisement

Nimrod-G and Virtual Lab Tools
for Data Intensive Computing on
Grid: Drug Design Case Study
Rajkumar Buyya
Melbourne, Australia
http://www.buyya.com/ecogrid
2
Contents
Introduction
Resource Management challenges
Nimrod-G Toolkit
SPMD/Parameter-Study Creation Tools
Grid enabling Drug Design Application
Nimrod-G Grid Resource Broker
Scheduling Experiments on
World Wide Grid
Conclusions
Grid
Economy
Grid
3
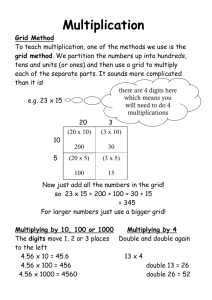
Scheduling
Economics
A typical Grid
environment and Players
Resource Broker
Application
Resource Broker
4
Grid Characteristics
Heterogeneous
Distributed
5
Resource Types: PC, WS, Clusters
Resource Architecture: CPU Arch, OS
Applications: CPU/IO/message intensive
Users and Owners Requirements
Access Price: different for different users, resources and time.
Availability: varies from time to time.
Resources
Ownership
Users
Each have their own (private) policies and objectives.
Very much similar to heterogeneity and decentralization that is
present in “human economies” (democratic and capitalist world).
Hence, we propose the use of “economics” as a metaphor for resource
management and scheduling. It regulates supply and demand for
resources and offers incentive for resource owners for contributing
resources to the Grid.
Grid Tools for Handling
Computational Economy
Security
Uniform Access System Management
Resource Discovery
6
Resource Allocation
& Scheduling
Application Development
Data locality
Network Management
Nimrod-G: Grid Resource Broker
A resource broker for managing, steering, and
executing task farming (parametric sweep/SPMD
model) applications on Grid based on deadline and
computational economy.
Based on users’ QoS requirements, our Broker
dynamically leases services at runtime depending on
their quality, cost, and availability.
Key Features
7
A single window to manage & control experiment
Persistent and Programmable Task Farming Engine
Resource Discovery
Resource Trading
Scheduling & Predications
Generic Dispatcher & Grid Agents
Transportation of data & results
Steering & data management
Accounting
Parametric Processing
Parameters
Age
23
23
28
28
19
10
-4000000
Hair
Clean
Beard
Goatee
Clean
Moustache
Clean
Too much
Multiple Runs
Same Program
Multiple Data
Magic Engine for
Manufacturing Humans!
8Courtesy: Anand Natrajan, University of Virginia
Killer Application for the Grid!
Sample P-Sweep Applications
Bioinformatics:
Drug Design / Protein
Modelling
Sensitivity
experiments
on smog formation
Ecological Modelling:
Combinatorial
Control Strategies
Optimization:
for Cattle Tick
Meta-heuristic
Data Mining
parameter estimation
Computer Graphics:
Ray Tracing
High Energy
Physics:
Searching for
Rare Events
Electronic CAD:
Field Programmable
Gate Arrays
VLSI Design:
Finance:
SPICE Simulations
Investment Risk Analysis
Civil Engineering:
Building Design
Automobile:
Crash Simulation
9
Network Simulation
Aerospace:
Wing Design
astrophysics
Virtual Drug Design: Data
Intensive Computing on Grid
A Virtual Laboratory for
“Molecular Modelling for Drug
Design” on Peer-to-Peer Grid.
It provides tools for
examining millions of chemical
compounds (molecules) in the
Protein Data Bank (PDB) to
identify those having potential
use in drug design.
In collaboration with:
10
http://www.csse.monash.edu.au/~rajkumar/vlab
Kim Branson,
Structural Biology,
Walter and Eliza Hall
Institute (WEHI)
Dock input file
score_ligand
minimize_ligand
multiple_ligands
random_seed
anchor_search
torsion_drive
clash_overlap
conformation_cutoff_factor
torsion_minimize
match_receptor_sites
random_search
. . . . . .
. . . . . .
maximum_cycles
ligand_atom_file
receptor_site_file
score_grid_prefix
vdw_definition_file
chemical_definition_file
chemical_score_file
flex_definition_file
flex_drive_file
ligand_contact_file
ligand_chemical_file
ligand_energy_file
11
yes
yes
no
7
no
yes
0.5
3
yes
no
yes
1
S_1.mol2
ece.sph
ece
parameter/vdw.defn
parameter/chem.defn
parameter/chem_score.tbl
parameter/flex.defn
parameter/flex_drive.tbl
dock_cnt.mol2
dock_chm.mol2
dock_nrg.mol2
Molecule to
be screened
Parameterize Dock input file
(use Nimrod Tools: GUI/language)
12
score_ligand
minimize_ligand
multiple_ligands
random_seed
anchor_search
torsion_drive
clash_overlap
conformation_cutoff_factor
torsion_minimize
match_receptor_sites
random_search
. . . . . .
. . . . . .
maximum_cycles
ligand_atom_file
receptor_site_file
score_grid_prefix
vdw_definition_file
chemical_definition_file
chemical_score_file
flex_definition_file
flex_drive_file
ligand_contact_file
ligand_chemical_file
ligand_energy_file
$score_ligand
$minimize_ligand
$multiple_ligands
$random_seed
$anchor_search
$torsion_drive
$clash_overlap
$conformation_cutoff_factor
$torsion_minimize
$match_receptor_sites
$random_search
Molecule to be
screened
$maximum_cycles
${ligand_number}.mol2
$HOME/dock_inputs/${receptor_site_file}
$HOME/dock_inputs/${score_grid_prefix}
vdw.defn
chem.defn
chem_score.tbl
flex.defn
flex_drive.tbl
dock_cnt.mol2
dock_chm.mol2
dock_nrg.mol2
Create Dock PlanFile
1. Define Variable and their value
parameter database_name label "database_name" text select oneof "aldrich"
"maybridge" "maybridge_300" "asinex_egc" "asinex_epc" "asinex_pre"
"available_chemicals_directory" "inter_bioscreen_s"
"inter_bioscreen_n" "inter_bioscreen_n_300" "inter_bioscreen_n_500"
"biomolecular_research_institute" "molecular_science"
"molecular_diversity_preservation" "national_cancer_institute"
"IGF_HITS" "aldrich_300" "molecular_science_500" "APP" "ECE" default
"aldrich_300";
parameter score_ligand text default "yes";
parameter minimize_ligand text default "yes";
parameter multiple_ligands text default "no";
parameter random_seed integer default 7;
parameter anchor_search text default "no";
parameter torsion_drive text default "yes";
parameter clash_overlap float default 0.5;
parameter conformation_cutoff_factor integer default 5;
parameter torsion_minimize text default "yes";
parameter match_receptor_sites text default "no";
parameter random_search text default "yes";
. . . . . .
. . . . . .
parameter maximum_cycles integer default 1;
parameter receptor_site_file text default "ece.sph";
parameter score_grid_prefix text default "ece";
parameter ligand_number integer range from 1 to 2000 step 1;
Molecules to be
screened
13
Create Dock PlanFile
2. Define Task that jobs need to do
task nodestart
copy ./parameter/vdw.defn node:.
copy ./parameter/chem.defn node:.
copy ./parameter/chem_score.tbl node:.
copy ./parameter/flex.defn node:.
copy ./parameter/flex_drive.tbl node:.
copy ./dock_inputs/get_molecule node:.
copy ./dock_inputs/dock_base node:.
endtask
task main
node:substitute dock_base dock_run
node:substitute get_molecule get_molecule_fetch
node:execute sh ./get_molecule_fetch
node:execute $HOME/bin/dock.$OS -i dock_run -o dock_out
copy node:dock_out ./results/dock_out.$jobname
copy node:dock_cnt.mol2 ./results/dock_cnt.mol2.$jobname
copy node:dock_chm.mol2 ./results/dock_chm.mol2.$jobname
copy node:dock_nrg.mol2 ./results/dock_nrg.mol2.$jobname
endtask
14
Use Nimrod-G
90
80
70
60
50
40
30
20
10
0
East
West
North
South
1st Qtr 2nd Qtr 3rd Qtr 4th Qtr
15
Submit & Play!
A Nimrod/G
Monitor
Cost
Deadline
66 Arlington
Alexandria
Legion hosts
She na nd o a h
Rive r
64
64
81
Ra p p a ha n no c k Po to m a c
Rive r
Rive r
Ja m e s
Rive r
Roanoke
Richmond
Ap p o m a to x
Rive r
Hampton
Norfolk
Virginia Beach
Portsmouth Chesapeake
Newport News
77
VIRGINIA
85
Globus Hosts
Bezek is in both
Globus and Legion Domains
16
Adaptive Scheduling Algorithms
Adaptive Scheduling
Algorithms
Time Minimisation
Cost Minimisation
None Minimisation
Execution Time
(not beyond deadline)
Minimise
Limited by deadline
Limited by deadline
Discover Establish
Resources
Rates
Distribute Jobs
17
Compose &
Schedule
Execution Cost
(not beyond budget)
Limited by budget
Minimise
Limited by budget
Discover
More
Resources
Evaluate &
Reschedule
Meet requirements ? Remaining
Jobs, Deadline, & Budget ?
Scheduling Experiment
on World Wide Grid Testbed
WW Grid
Cardiff/UK
Portsmoth/UK
TI-Tech/Tokyo
ETL/Tsukuba
AIST/Tsukuba
ANL/Chicago
USC-ISC/LA
UTK/Tennessee
UVa/Virginia
Dartmouth/NH
BU/Boston
Santiago/Chile
18
EUROPE:
ZIB/Germany
PC2/Germany
AEI/Germany
Lecce/Italy
CNR/Italy
Calabria/Italy
Pozman/Poland
Lund/Sweden
CERN/Swiss
Kasetsart/Bangkok
Monash/Melbourne
VPAC/Melbourne
Deadline and Budget Constrained
Scheduling Experiment
Workload:
Deadline: 2 hrs. and budget: 396000 units
Strategy: minimise time / cost
Execution Cost with cost optimisation
19
165 jobs, each need 5 minute of CPU time
Optimise Cost: 115200 (G$) (finished in 2hrs.)
Optimise Time: 237000 (G$) (finished in 1.25 hr.)
In this experiment: Time-optimised scheduling run
costs double that of Cost-optimised.
Users can now trade-off between Time Vs. Cost.
World Wide Grid (WWG)
WW Grid
Australia
North America
ANL: SGI/Sun/SP2
USC-ISI: SGI
UVa: Linux Cluster
UD: Linux cluster
UTK: Linux cluster
Monash Uni.:
Nimrod/G
Linux cluster
Globus+Legion
GRACE_TS
Solaris WS
Globus/Legion
GRACE_TS
Asia/Japan
WW Grid
Internet
Tokyo I-Tech.:
ETL, Tuskuba
Linux cluster
Globus +
GRACE_TS
Chile: Cluster
20
Globus +
GRACE_TS
South America
Europe
ZIB/FUB: T3E/Mosix
Cardiff: Sun E6500
Paderborn: HPCLine
Lecce: Compaq SC
CNR: Cluster
Calabria: Cluster
CERN: Cluster
Pozman: SGI/SP2
Globus +
GRACE_TS
Resources Selected &
Price/CPU-sec.
21
Resource &
Location
Grid services
& Fabric
Cost/CPU
sec. or
unit
No. of Jobs Executed
Linux Cluster-Monash,
Melbourne, Australia
Globus, GTS, Condor
2
64
153
Linux-Prosecco-CNR,
Pisa, Italy
Globus, GTS, Fork
3
7
1
Linux-Barbera-CNR,
Pisa, Italy
Globus, GTS, Fork
4
6
1
Solaris/Ultas2
TITech, Tokyo, Japan
Globus, GTS, Fork
3
9
1
SGI-ISI, LA, US
Globus, GTS, Fork
8
37
5
Sun-ANL, Chicago,US
Globus, GTS, Fork
7
42
4
Time_Opt
Cost_Opt
Total Experiment Cost (G$)
237000
115200
Time to Complete Exp. (Min.)
70
119
DBC Scheduling for Time
Optimization
Condor-Monash
Linux-Prosecco-CNR
Linux-Barbera-CNR
Solaris/Ultas2-TITech
SGI-ISI
Sun-ANL
12
No. of Tasks in Execution
10
8
6
4
2
22
Time (in Minute)
68
72
60
64
52
56
44
48
36
40
28
32
20
24
16
8
12
4
0
0
DBC Scheduling for Cost
Optimization
Condor-Monash
Linux-Prosecco-CNR
Linux-Barbera-CNR
Solaris/Ultas2-TITech
SGI-ISI
Sun-ANL
14
No. of Tasks in Execution
12
10
8
6
4
2
Time (in Minute)
23
10
2
10
8
11
4
96
90
84
78
72
66
60
54
48
42
36
30
24
18
6
12
0
0
Conclusions
24
P2P and Grid Computing is emerging as a next generation
computing platform for solving large scale problems through
sharing of geographically distributed resources.
Resource management is a complex undertaking as systems need
to be adaptive, scalable, competitive,…, and driven by QoS.
We proposed a framework based on “computational economies” and
discussed several economic models for resource allocation and for
regulating supply-and-demand for resources.
Scheduling experiments on World Wide Grid demonstrate our
Nimrod-G broker ability to dynamically lease or rent services at
runtime based on their quality, cost, and availability depending on
consumers QoS requirements.
Easy to use tools for composing applications to run on Grid are
essential to attracting and getting application community on board.
Economics paradigm for QoS driven resource management is
essential to push P2P/Grids into mainstream computing!
Download Software & Information
Nimrod & Parameteric Computing:
Economy Grid & Nimrod/G:
http://www.buyya.com/vlab/
Grid Simulation (GridSim) Toolkit (Java based):
http://www.buyya.com/ecogrid/
Virtual Laboratory/Virtual Drug Design:
http://www.csse.monash.edu.au/~davida/nimrod/
http://www.buyya.com/gridsim/
World Wide Grid (WWG) testbed:
http://www.buyya.com/ecogrid/wwg/
Looking for new volunteers to grow
25
Please contact me to barter your & our machines!
Want to build on our work/collaborate:
Talk to me now or email: rajkumar@csse.monash.edu.au