V h
advertisement
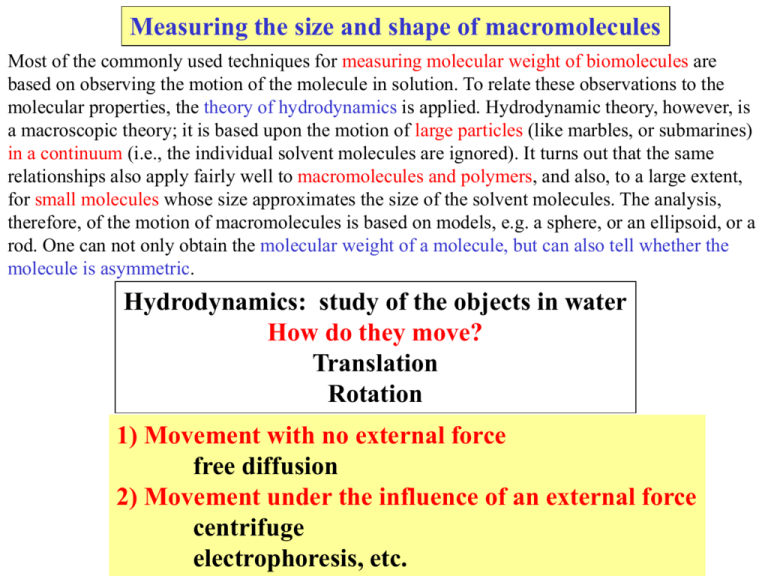
Measuring the size and shape of macromolecules Most of the commonly used techniques for measuring molecular weight of biomolecules are based on observing the motion of the molecule in solution. To relate these observations to the molecular properties, the theory of hydrodynamics is applied. Hydrodynamic theory, however, is a macroscopic theory; it is based upon the motion of large particles (like marbles, or submarines) in a continuum (i.e., the individual solvent molecules are ignored). It turns out that the same relationships also apply fairly well to macromolecules and polymers, and also, to a large extent, for small molecules whose size approximates the size of the solvent molecules. The analysis, therefore, of the motion of macromolecules is based on models, e.g. a sphere, or an ellipsoid, or a rod. One can not only obtain the molecular weight of a molecule, but can also tell whether the molecule is asymmetric. Hydrodynamics: study of the objects in water How do they move? Translation Rotation 1) Movement with no external force free diffusion 2) Movement under the influence of an external force centrifuge electrophoresis, etc. Hydrodynamics techniques measure the frictional resistance of the moving macromolecule in solution: 1. Intrinsic viscosity the influence of the object on the solution properties 2. Free translational diffusion 3. Centrifugation/sedimentation 4. Electrophoresis 5. Gel filtration chromatography 6. Free rotational diffusion/fluorescence anisotropy What do we learn here? 1. We will be examining the basis for several techniques that rely on the movement of macromolecules in solution to reveal information about their size and shape, or separate molecules based on these differences. 2. We will discuss the results of frictional energy dissipation due to the solvent moving about a macromolecule (referred to as a “particle”), which changes the solution viscosity, and the affect of this frictional drag on the diffusion of macromolecules in solution. 3. We will consider methods that have become routine laboratory techniques such as SDS-polyacrylamide electrophoresis, gel filtration chromatography, and sedimentation. Hydrodynamics Measurements of the motion of macromolecules in solution form the basis of most methods used to determine molecular size , density , shape , molecular weight Equations based on the behavior of macroscopic objects in water can be used successfully to analyze molecular behavior in solution sphere ellipsoid (prolate, oblate) disk rod etc. Techniques Free Motion Viscosity translational diffusion rotational diffusion Mass Transport sedimentation electrophoresis gel filtration The hydrodynamic particle hydrodynamically bound water shear plane: frictional resistance to motion depends on size shape A macromolecule in solution is hydrated and when it moves the shear plane that defines the boundary of what is moving includes the associated water. For a globular protein the water of hydration is approximately one layer of water. Recall that this is very rapidly exchanging with the bulk solvent water, but nevertheless this associated water of hydration defines the actual hydrodynamic particle. Hydrodynamic Parameters 1.: Specific Volume: (inverse of density) volume Vi gram If there are g1 and g2 grams of solvent and solute then Volume = V1g1 + V2g2 total volume: vol gram Consider a two component system; solvent (1) and solute (2) x grams The specific volume is the inverse of the density of an anhydrous macromolecule. This quantity which is more relevant from a thermodynamic viewpoint. Proteins: V2 ~ 0.7 - 0.75 mL/g Na+ DNA: V2 ~ 0.5 - 0.53 mL/g Hydrodynamic Parameters 2. Partial Specific Volume: change in volume per gram under specific conditions V Vi = ( ) gi T,P,gj Volume = V1g1 + V2g2 Gibbs - Duham relationship We will assume that specific volume is approximately equal to the partial specific volume: Vi Vi Proteins: VP ~ 0.7 - 0.75 mL/g Na+ DNA: VDNA ~ 0.5 - 0.53 mL/g For proteins, the value of the specific volume can be accurately predicted by summing the specific volumes of the component individual amino acids. Note that V2 is the inverse of the density of the anhydrous macromolecule. This can be measured directly by observing the volume change upon dissolving the macromolecule, or the density change of the solution. The bar on top of the V denotes volume per gram. Hydrodynamic Parameters 3. Effective mass This is the molecular mass minus the mass of the displaced solvent (the buoyant factor). Take into account buoyancy by subtracting the mass of the displaced water [mass - (mass of displaced water)] [m - m(V2) = m (1 - V2)] M (1 - V2) N mV2 = volume of particle mV2 = mass of displaced solvent solvent density V2 = Vol of the molecule g m = (M/N) mass of particle N = Avogadro number M = molecular weight Hydrodynamic Parameters 4. Hydration 1= gH2O gprotein mh = m + m 1 mh = M ( 1 + 1) N Vh = M ( V2 + 1 V1) N Vh = (V2 + 1 V1) Mass of hydrated protein Volume of hydrated protein Volume of hydrated protein per gram of protein Problems of hydrodynamic measurements in general: the properties depend on too many parameters! 1) size 2) shape 3) solvation Some solutions to obtain useful information: 1 Use more than one property and eliminate one unknown. Example: sedimentation (So) and diffusion (D) can eliminate shape factor 2 Work under denaturing conditions to eliminate empirically the shape factor and solvation factors. These are constants for both standards and the unknown sample - then find Molecular weight. Examples: 1) gel filtration of proteins in GuHCl; 2) SDS gels. 3 If Molecular weight is known: compare results with predictions based on an unhydrated sphere of mass M. Then use judgement to explain deviations on the basis of shape or hydration. Behavior of limiting forms – e.g; rods, spheres- are calculated for comparisons All hydrodynamic measurements yield a Stokes’ radius. In general, the results of hydrodynamic measurements are interpreted in terms of the Stokes’ radius, Rs, of the particle. The Stokes’ radius is the radius of a spherical particle which would give the measured hydrodynamic property (intrinsic viscosity, diffusion, sedimentation, electrophoretic mobility or gel filtration elution). If the molecular weight (M) of the protein (or particle) is known along with its density, then the radius can be readily calculated if it is assumed that the protein is spherical. This is called the minimal radius, Rmin, which refers to the unhydrated protein. One can then compare the Stokes radius (Rs) to the calculated Rmin. If they are similar, then it is likely that the protein really is a sphere, since the calculated and experimental quantities are in agreement. If they are not similar, then one of the assumptions used to calculate the value of Rmin must be incorrect. The assumptions are that the protein (or particle) is not hydrated and that the particle is spherical. The inclusion of the water of hydration results in a modest change in particle size for a compact particle such as a globular protein. On the other hand, as we shall see, a highly asymmetric particle (e.g., very elongated) can cause a major discrepancy. Essentially, a highly asymmetric molecule behaves as a sphere with a size much larger than the value of Rmin. There is a large frictional drag on the movement of a very asymmetric molecule in solution or through a gel matrix. The overall strategy is summarized in the next slide. Next we consider: 1 What effect does the interaction of the hydrodynamic particle and the solvent have on solution properties? viscosity 2 What effect does the particle-solvent interaction have on the motion of the particle? translational motion - free - in a force field rotational motion Viscosity Frictional interactions between “layers” of solution results in energy dissipation u (velocity) dy u + du A Ff du • A du =•( )•A dy dy (shearing force) coefficient of viscosity cgs units Poise is related to the amount of energy dissipated per unit volume per unit time Many ways to measure solution viscosity capillary viscometer: measures the rate of flow of solution through a capillary with a pressure drop P Effect of macromolecules on viscosity: only 2 parameters plus solute =1+ • o solvent alone fraction of volume occupied by “particles” “shape” factor (also called the Simha factor) 20 Shape factor For a sphere: = 2.5 Shape factor is very large for highly asymmetric molecules 10 2.5 1 5 10 15 axial ratio = a/b b a viscosity effects are very large for very elongated polymers (large ) (e.g. actin) or molecules that “occupy” large volume (large ) (e.g. DNA) Relative viscosity and Specific viscosity o = r relative viscosity = 1 + - o = sp specific viscosity o sp = r - 1 = = shape factor = fraction of solution volume occupied by solvent particles c = g / mL solvent But = Vh • c so sp = • Vh c Vh = volume per gram of protein occupied by solvated particle ideally, the increase in specific viscosity per gram of solute should be a constant ( • Vh ). Often it is not due to “non-ideal” behavior Shape factor vol/g of protein of hydrated particle Non-ideal case (i.e.,reality): sp = • Vh + (const.) • c + .... c sp c (ideal) c Extrapolate measurement to infinite dilution: Intrinsic viscosity lim (sp /c) = [] limit at low concentration (intercept in graph above) C0 Intrinsic viscosity: [], units = mL / g [] = • Vh Intrinsic viscosity is dependent only on size and properties of the isolated macromolecule. Intrinsic Viscosities of Proteins in Aqueous Salt Solution Molecular Dimensions of Proteins from Intrinsic Viscosities Intrinsic viscosity [] = • Vh monomer = 2.5 [] = • Vh increases polymer if = 25 then [] is 10x larger [] = • Vh if we assume a spherical shape, = 2.5, we will calculate an incorrect and very large value for Vh The Intrinsic Viscosity of Flexible Polymers (DNA, Polysaccharides, etc.) For flexible polymers, several theories have been offered to compute the frictional interactions with the solvent. It is not clear how one could decide what the value of ϕ should be for a large flexible polymer with lots of solvent within the polymer matrix. It turns out that most of the solvent within RG, the radius of gyration, from the center of mass must be relatively immobile so that the flexible polymer behaves similar to that expected for a compact sphere with R ≈ 0.8RG. Hence, the flexible polymer occupies effectively a much larger fraction of solution than with the same mass of a compact polymer. For a flexible polymer like DNA or denatured protein Behaves like compact particles with R 0. 8 x radius of gyration (radius of gyration measures mass distribution and can be obtained from light scattering) example: [] 4 mL / g for a native, globular protein But for a random coil, if there is ~ 100 g “bound” solvent per gram solute Vh V2 + 100 100 mL/ g [] = 2.5 • Vh very large “coil” Viscosity : examples I. Hemocyanin (M = 106): native vs GuHCl denatured sp C 60 GuHCl denatured 40 [] = • Vh = 2.5 Vh coiled 20 0 cm3 g Native 0 5 compact 10 15 C (mg / mL) of BSA For a native, globular protein, [η] ≈ 4 ml/g. [η ] for same protein denatured in Gu • HCl is about 50 ml/g. If the amount of water within the flexible polymer is about 100 g per gram of solute (δ1=100), then Vh ≈ 100 ml/g. II. T7 DNA (from phage T7, double strand): salt dependence 3000 [sp] C expanded coil 2000 1000 cm3 g 0 0 0.01 0.1 log [NaCl] 1.0 Note the huge values of [η ] at low ionic strengths. This results from the expansion of the DNA due to charge repulsion of the phosphates. A typical value of [η] for double strand DNA at 0.2 M NaCl is about 100 ml/g. Stokes Radius Radius of the sphere which has the hydrodynamic properties consistent with the hydrodynamic measurement [] = • Vh 1. measure [] = 2.5 • Vh 2. assume a sphere 3. calculate Stokes radius 4/3 Rs3 M/N volume of the molecule (spherical) mass of the molecule Solve for the Stokes radius: Rs = [ M 3 • Vh]1/3 N 4 where Vh = []/2.5 MINIMUM RADIUS The radius expected if the molecule is an anhydrous sphere [4/3 Rmin3] V2 = (M/N) Rmin = [ M 3 1/3 V ] 2 N 4 anhydrous sphere - point of reference CALCULATED Compare the Stokes radius (measured) to the “minimum radius” expected assuming the molecule is an anhydrous sphere Question: How close does the assumption of an anhydrous sphere come to explaining the value of Rs? If Rs/Rmin is not much larger than 1.0, then the assumption of the molecule being spherical is likely reasonable. Rs should be slightly larger than Rmin due to hydration If Rs/Rmin is much larger than 1.0, then either the molecule is not spherical ( >>2.5) or it is not compact (Vh>>V2) For an anhydrous particle: anhydrous vol/gram [] = • V2 for an anhydrous sphere: = 2.5 [] = 2.5 • V2 For real macromolecules, the intrinsic viscosity will vary from the above due to 1. correction due to hydration Vh (hydrated vol/g = V2 +H20) = 2.5 2. correction due to asymmetry > 2.5 Vh = V2 (anhydrous vol/g) or both hydration and asymmetry > 2.5 and Vh = V2 + H2O Comparison of intrinsic viscosity values for two proteins: 1. Ribonuclease: mol wt: 13, 683 V2 = 0.728 ml/g Rmin = 17 Å 2. Collagen: mol wt: 345,000 V2= 0.695 ml/g Rmin = 59 Å Interpreting Viscosity Data: Does a reasonable amount of hydration explain the measured value of []? protein [] mL/g Ribonuclease 3.3 Collagen 1150 maximum solvation maximum asymmetry Rs (Å) H2O (g/g) (a/b) 19.3 0.59 4.5 3.9 400 460 1660 175 [] = Vh = [4/3 Rs3] N M Stokes Radius Rs = ( Appropriate for collagen Rs/Rmin = 6.8 (400/59) 3 [] M 1/3 ) 4N Vh = V2 (anhydrous value) shape correction: > 2.5 hydration correction: Vh = V2 + H2O Appropriate for ribonuclease Rs/Rmin = 1.14 (19.3/17) 2.5 = 2.5





