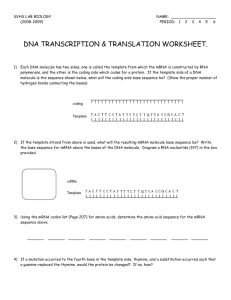
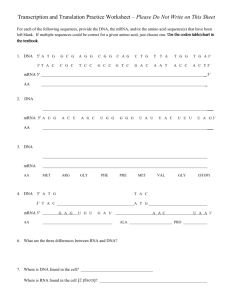
DNA

Chapter 12 and 13
Nucleic Acids, Protein Synthesis and Mutations
Central Dogma
DNA RNA Protein
1
Identifying the substance of Genes
• Influential Scientists:
• Griffith - Experimented with mice and bacteria that cause pneumonia and demonstrated TRANSFORMATION .
- Concluded that some FACTOR (gene) was responsible for the change.
• Avery - Followed up on Griffith’s experiment; wanted to know which molecule was important for transformation.
He then extracted the 4 macromolecules from these heat killed cells, all of which were considered to be possible candidates for the carriers of genetic information.
Treated each mixture with enzymes that destroyed the macromolecule and transformation still occurred.
He concluded that DNA is the source of genetic information.
Figure 12–2 Griffith’s
Section 12-1
Experiment
Heat-killed, disease-causing bacteria (smooth colonies)
Disease-causing bacteria (smooth colonies)
Harmless bacteria
Heat-killed, disease-
(rough colonies) causing bacteria
(smooth colonies)
Control
(no growth)
Harmless bacteria
(rough colonies)
Dies of pneumonia Lives Lives
Live, disease-causing bacteria (smooth colonies)
Dies of pneumonia
DNA
• Influential Scientists continued:
• Hershey and Chase -
Studied viruses – non-living particles that can infect living cells.
- Looked at bacteriophages – a virus that infects bacteria.
Supported the conclusion that genes were made of DNA.
DNA is the source of genetic information
Bacteriophage with phosphorus-32 in DNA
Phage infects bacterium
Radioactivity inside bacterium
Bacteriophage with sulfur-35 in protein coat
Phage infects bacterium
No radioactivity inside bacterium 4
Where is DNA found?
Inside the nucleus DNA is coiled into Chromosomes
5
Components of DNA
• Nucleotide (monomer)
– Deoxyribose sugar
– Phosphate group
– Nitrogen -containing base
• Adenine ( A )
• Guanine ( G )
• Cytosine ( C )
• Thymine ( T )
Purines
Pyrimidines
6
Components of DNA
• The structure or shape of DNA = Double Helix =
2 strands
– Watson and Crick (1953) – tried to assemble the structure.
– Rosalind Franklin (1952) – used a technique known as x-ray diffraction to create a picture.
• The x shape indicated DNA is twisted (helix) around two strands.
7
Components of DNA
• Complementary Base Pairing
– A↔T, G ↔C (Chargaff’s rule)
– Connected by covalent hydrogen bonds
DNA with
DNA Replication (duplication)
• Takes place in the nucleus (during the S phase)
• Result: 2 exact copies original DNA
DNA Polymerase
Helicase
Replication fork
9
DNA Replication (duplication)
• 1. Helicase unzips the double helix by breaking the hydrogen bonds forming a replication fork.
• 2. DNA polymerase adds the complimentary base pairs to each separated strand.
– DNA Polymerase also “ proofreads ” each new strand.
Helicase DNA Polymerase
Replication fork
10
DNA Replication, Accuracy & Repair
• Original: A-T-T-C-C-G
• Complement: TAAGGC
• Original: GCTAAG
• Complement:
• Original: CTACCA
• Complement:
• Original
– Strand A: GACCTA
– Strand B:
• DNA polymerase proofreads & repairs 1error/1Billion
11
• Nucleotide
– Ribose Sugar
Components of RNA
– Phosphate Group
– Nitrogen Base
• Adenine (A)
• Guanine (G)
• Cytosine (C)
• Uracil (U): not T
• Single Strand
• 3 Types:
– Messenger RNA (mRNA)
– Transfer RNA (tRNA)
– Ribosomal RNA (rRNA)
12
How to make RNA
Step 1 = Transcription : DNA RNA
Takes place in the nucleus
1.
DNA unwinds.
2.
RNA Polymerase binds to DNA promoter site (begin gene)
Gene Begins
RNA Polymerase
3. Add complementary RNA nucleotides (U↔A)
13
Transcription Continued
4. DNA termination sequence signals gene end
RNA Polymerase
5. RNA Polymerase releases DNA & RNA
RNA Strand
DNA Rewinds
14
Transcription
DNA makes RNA
15
1.
Carries instructions from
DNA to assemble amino acids into protein.
3 Types of RNA
3.
The site where proteins get assembled from mRNA.
2.
Carries the amino acids to the mRNA at the ribosome.
16
How to make Protein =
Translation
• Involves the decoding of mRNA and assembling a protein
• Proteins = polymers = macromolecule
– Monomer of protein = amino acid
– Polypeptides = sequence of amino acids
• Genetic code is read 3 letters at a time.
– Codon: every 3 base pairs in mRNA = an amino acid
• START Codon : starts translation- 1 codon only AUG
• STOP Codon : stops translation- 3 codons UAA, UAG, UGA
– Universal Codon -Amino Acid Code: p. 367
17
mRNA Codon & Codon Chart
• AUG =
– Methionine or start codon
• AAC = _________
Amino Acids
18
19
How to make Protein =
Translation
• tRNA
– In cytosol
– Binds specific amino acid to mRNA
– Anticodon :
• complement to mRNA codons
20
Translation: mRNA
Protein
1.
mRNA leaves nucleus
2.
Ribosome attaches to mRNA start codon
3.
mRNA codon pairs with tRNA anticodon delivering amino acid.
4.
Peptide bond forms between amino acids
21
Translation: mRNA
Protein
5.
mRNA stop codon signals end of translation. The ribosome releases the newly formed polypeptide.
6.
mRNA released & polypeptide complete
22
Nucleus
Translation Diagram
Polypeptide Chain Peptide Bond
Amino Acid tRNA mRNA Codon
Ribosome
Anticodon
23
Overview
DNA RNA Protein
Transcription
Translation
24
Mutations
• Mutations – are heritable changes in genetic info.
• Occurs in only 2 types of cells
– Sex-cell (germ-cell) mutations: gametes affect offspring
– Somatic mutations: body cells affect individual
• 2 categories of mutations
– Gene mutations produce change in a single gene
– Chromosomal mutations produce change in a whole chromosome.
25
Gene Mutations
A.K.A. Point Mutations (3 types)
1.
Substitution 1 nitrogen base gets substituted by another nitrogen base; this results in a new codon
– Sickle Cell Anemia : substitute A for T
26
Gene Mutations (con’t)
2.
Nucleotide deletions & insertions
– One base is inserted or removed from the sequence.
Causes Frame-shift mutations
– Changes amino acid sequence
Deletion
Insertion
Deletion
27
Chromosomal Mutations (5 types)
• Deletion: lose portion
• Duplication: gain extra portion
• Inversion: segment reverses
• Translocation: transfer segment to non-homologous
• Nondisjunction: gamete gets extra or less chromosome (Down Syndrome- Trisomy 21)
28
Chromosome Mutations Diagrams
29
Genetic Traits & Disorders
• Disorders due to nondisjunction
– Nondisjunction: gametes have 1 more or less chromosome (pairs don’t segregate)
– Monosomy: 45 chromosomes
• Turner’s syndrome: XO
– Trisomy: 47 chromosomes
• Down Syndrome: trisomy-21
• Kleinfelter’s syndrome: XXY
• Patau syndrome: trisomy-13
• Edward’s syndrome: trisomy-18
30