PPPP-Cooper-5-16-13... - Buffalo Ontology Site
advertisement

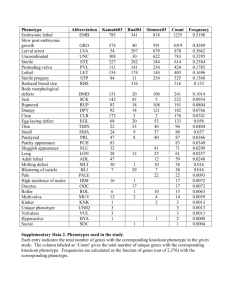
Plant Phenotype Pilot Project The Issue: Traditional free text phenotype descriptions are inadequate for large-scale computerized comparative analyses AIM: To use ontologies in express and analyze plant phenotypes from multiple species Phenotype Ontology Research Coordination Network http://www.phenotypercn.org/ 4 Working Groups: Vertebrates Arthropods Plants Informatics Many fields of biology represented: Systematics Evolutionary biology Genetics/developmental biology Ecology Paleontology … Unifying ideas: Shared Ontologies Shared tools and methods Best practices Community outreach Challenges of managing phenotype data • Extremely diverse data type (can range from expression profile to behavior) • Can be associated to individuals, populations or species • Different levels (summary, measurement data) • Can be comparative (mutant vs. wild type) or absolute (days to flowering of a cultivar) • Data integration - needs extensive connections to other types of data (seed stocks, genes, experimental methods, publications) • Database schema and interface design Data representation - how to represent the data in a consistent way across experiments, research communities and species Data accessibility – how do we get data out of literature and into the database? Collection of phenotype data- Who is involved? Species Glycine max Solanum lycopersicum Medicago truncatula Zea mays Oryza sativa Arabidopsis thaliana Genes included in project set 233 74 443 324 138 2400 Source SoyBase SGN LIS MaizeGDB PO/Gramene/Oryzabase Lloyd and Meinke 2012 Pilot Project - limited scope: • Mutant phenotypes (not natural variants) • Emphasis on visual and morphological (no gene expression patterns) • Summary data (not phenotype measurements) Phenotype Data: Phenotypic measurement Genotype measured Leaves are 1 cm wide Growth conditions Control treatment Image Reference genotype Experimental treatment Data collection method Statistical method Data interpretation – preferably done by experimenter Phenotype Summary: Mutant yfg1-1 has narrow leaves and flowers early in short days Why use ontologies? • Supplement, not replacement, for free text • Provides standardized vocabulary – Dwarf, short stature, small plant, reduced height are different ways of expressing the same idea • Provides relationships among terms – Vascular leaf is_a type of leaf – Leaf abscission zone part_of leaf – Leaf develops_from leaf primordium • Makes computational approaches possible – Searches – Categorization – Network analysis, semantic similarity Outline of Pilot Project Existing phenotype datasets: Phenotypes of mutant loci, QTL Existing reference ontologies Phenotypes of cloned genes Plant Ontology Gene Ontology PATO ChEBI Plant EO Consistent and thorough set of ontology annotations Ontology statements Semantic similarity computational analysis Phenotypes and Ontologies: From an ontological perspective, a phenotype is a combination of an entity and a quality that inheres in that entity Phenotype name adherent leaf notched petal high yield increased water loss inheres in Entity juvenile vascular leaf petal seed transpiration Quality fused lobed increased mass increased rate Phenotypes may also consist of two entities and a relationship between them: Entity 1 juvenile vascular leaf gynoecium Relationship* fused with basal to *in PATO, the relationship is called a “relational quality” Entity 2 stem perianth Examples of mutant phenotypes shared across species: Dwarf plants Rolled leaves Examples Description of Mutant Phenotype Atomized Phenotype statements dwarf Dwarf with profuse slender tillers, small panicles PO: shoot system profuse tillers PO: whole plant PO: basal axillary shoot system small panicles PO: inflorescence slender tillers Delayed flowering; Reduction in total chlorophyll Quality (PATO) Entity GO: flowering decreased height has extra parts of type (basal axillary shoot system) slender decreased size delayed ChEBI: chlorophyll decreased concentration Next steps: • Data analysis • Clustering of genes into pathways • Degree of correlation between sequence and phenotype • Computational prediction of gene candidates for uncloned mutant genes and QTL • Apply lessons learned • Is the data set big enough? • Are the ontologies complete enough? • Is our annotation consistency good enough? • Better analysis methods? Future Possibilities with cROP • Expansion to use Protein Ontology Plant Ontology ChEBI Gene Ontology Ontology statements Plant EO PATO PRO Acknowledgements USDA-ARS-CICGRU: Oklahoma State University: David Meinke Steven Cannon, Scott Kalberer Michigan State University: Johnny Lloyd Carolyn Lawrence, Lisa Harper U. Of Nottingham Sean May Rex Nelson, David Grant Boyce Thompson Institute: Lukas Mueller (SGN) Naama Menda (SGN) University of Arizona: Ramona Walls (PO / iPlant) Oregon State University: Laurel Cooper Pankaj Jaiswal Laura Moore George Gkoutos (University of Aberystwyth) Anika Oellrich (EBI) Funding: NSF - Phenotype Ontology Research Coordination Network (RCN) Ontolog y