Biology 6B Laboratory Report II
advertisement

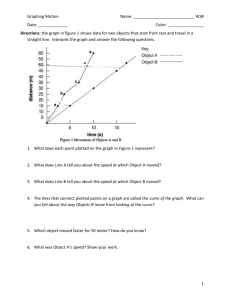
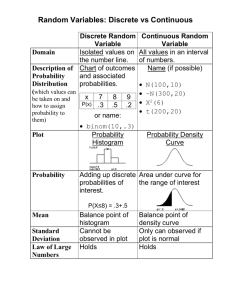
Cloning Worksheet Winter 2011 Producing a Standard Curve to Determine DNA Fragment Sizes Measuring Distance Traveled on Agarose Gel • Using a ruler and gel photograph, measure how far each DNA fragment traveled from its loading well • Choose a consistent starting point, either the middle or edge of the well • Measure from the starting point to the middle of each DNA band in metric units • Plot the log of the molecular weight (in base pairs) versus the distances traveled for the standard “ruler” fragments • Put a best fit line through these points • Determine the sizes of the PCR products, plasmids and inserts by interpolating from the standard curve of the ruler fragments • You may use semi-log paper to plot the line by hand or complete the analysis in Excel Measuring Distance Traveled Measure from the well to the middle of each band. Plot the standard curve with these values. Measure the distance for plasmids and fragments. Use the standard curve to find their sizes. Standard Semi-Log Plot Molecular Weight (base pairs) 100,000 10000 1000 100 Distance Traveled (cm) Standard Semi-Log Plot Distance Traveled (cm) 1.8 Distances 1.9 measured 2.0 from gel photograph 2.1 2.25 2.5 2.8 3.15 3.75 4.65 MW (bp) 5000 4500 4000 3500 3000 2500 2000 1500 1000 500 Standard sizes of the “ruler” fragments 100,000 Omit data that doesn’t show a linear relationship. 10000 X XX X X Use the best fit line. X X X 1000 X X 100 1 2 3 4 5 100,000 Finding the size of an unknown fragment 10000 X XX X X MW is ~900 bp X X X 1000 X X Fragment at 3.8 cm 100 1 2 3 4 5 Before plotting the standard curve, use the Excel function =LOG10(x) to convert MW numbers to log values Log Molecular Weight (bp) Using Excel to Estimate Fragment Size 4 3.5 3 2.5 Log MW 2 Linear (Log MW) 1.5 1 0.5 0 0 2 4 6 Distance Traveled (cm ) Trendline equation: y = -0.3442x + 4.2808 Solve for y using x = 3.8: y=2.97, Raise 10 to the y power: 102.97 = 939 base pairs