Gene Lists - HSLS - University of Pittsburgh
advertisement

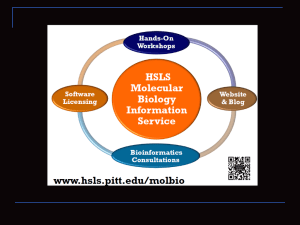
Pathway Informatics 6th July, 2015 Ansuman Chattopadhyay, PhD Head, Molecular Biology Information Services Health Sciences Library System University of Pittsburgh ansuman@pitt.edu Biological Pathway Map http://www.hsls.pitt.edu/molbio Pathway Informatics L i s t o f G e n e s Protein-Protein Interaction Database P a t h w a y M a p http://www.hsls.pitt.edu/molbio Gene list to pathways Gene List Biological Interaction Network B-factor, properdin APEX nuclease (multifunctional DNA repair enzyme) 1 ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase) proliferating cell nuclear antigen heterogeneous nuclear ribonucleoprotein A1 fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) three prime repair exonuclease 2 ligase I, DNA, ATP-dependent flap structure-specific endonuclease 1 cyclin A2 cyclin-dependent kinase inhibitor 1A (P21) checkpoint kinase 1 homolog (S. pombe) replication protein A1, 70kDa Werner syndrome exonuclease 1 uracil-DNA glycosylase 2 Biological Table Name Time 0 Time 15 mins Time 1 hr Time 5 hr IL6 35.45% 36.46% 95.34% 13.91% PDGFRA 32.42% 45.43% 30.99% 56.62% DCN 21.42% 1.29% 33.13% 41.40% ZFP36L1 48.17% 47.56% 84.31% 6.72% DEGS 46.41% 90.30% 52.14% 31.61% GNB2 42.37% 98.25% 25.28% 53.26% ATRN 93.42% 75.90% 1.56% 82.61% PDK4 88.18% 38.03% 95.95% 5.78% MAPK1 20.88% 64.99% 43.18% 52.77% THBS1 99.61% 54.35% 85.28% 85.69% CD68 9.35% 44.81% 69.03% 64.88% DIAPH2 54.16% 34.47% 63.26% 33.46% Sep6 30.12% 48.83% 28.57% 76.28% BFAR 10.18% 7.65% 60.30% 42.66% CCRK 79.03% 19.54% 96.15% 88.79% MT1E 49.07% 23.60% 93.13% 15.83% http://www.hsls.pitt.edu/molbio Topics Databases Biological Pathway Maps Protein-Protein Interaction High-throughput gene expression (Microarray) Database http://www.hsls.pitt.edu/molbio Topics Software Create a gene list: Geo2R, NextBio Pathway Analysis : NIH DAVID, Ingenuity Metacore, http://www.hsls.pitt.edu/molbio IPA, Pathway Databases http://www.hsls.pitt.edu/molbio Biological Pathways Metabolic Signaling http://www.hsls.pitt.edu/molbio Biological Pathway Databases KEGG Ingenuity IPA GeneGo Metacore Millipore Pathways (Metacore) Sigma Your Favorite Genes (IPA) Life Technologies GeneAssist Pathway Commons http://www.hsls.pitt.edu/molbio Databases-Pathways NCBI Biosystems KEGG http://www.ncbi.nlm.nih.gov/biosystems Pathway Commons: http://www.pathwaycommons.org/pc/ YFG – Sigma Aldrich: Your Favorite Gene Millipore Pathways: http://www.millipore.com/pathways/pw/pathways Applied BioSystems: http://www5.appliedbiosystems.com/tools/pathway/ http://www.hsls.pitt.edu/molbio PPI Databases http://www.hsls.pitt.edu/molbio What proteins interact with my favorite protein? http://www.hsls.pitt.edu/molbio PPI Databases BioGRID STRING http://www.hsls.pitt.edu/molbio - Retrieve interacting partners of a protein of your interest -What proteins interact with human EGFR? Resources BioGrid: http://thebiogrid.org/ STRING: http://string-db.org/ Link to the video tutorial: http://media.hsls.pitt.edu/media/molbiovideos/ppi.swf BioGrid http://www.hsls.pitt.edu/molbio BioGrid Search Result Page http://www.hsls.pitt.edu/molbio STRING - Known and Predicted ProteinProtein Interactions An ultimate resource for finding both predicted and experimental verified PPI Not limited to human, mouse and rat Developed at CPR, EMBL, SIB, KU, TUD and UZH http://www.hsls.pitt.edu/molbio STRING http://www.hsls.pitt.edu/molbio Gene Lists http://www.hsls.pitt.edu/molbio Gene Lists Microarrays Protein arrays CHIP-chip SNP arrays RNA Seq Literature Search http://www.hsls.pitt.edu/molbio Gene Expression Databases NCBI • Gene Expression Omnibus (GEO) EBI • ArrayExpress http://www.hsls.pitt.edu/molbio Gene Expression Database http://www.hsls.pitt.edu/molbio Use of GEO Data http://goo.gl/yD2hR http://www.hsls.pitt.edu/molbio Generate a gene list http://www.hsls.pitt.edu/molbio Pathway Analysis Control Treated A549 cell + Ethanol (25mmol/L) 48 hr A549 cell + Resveratrol (25mmol/L) 48 hr http://www.hsls.pitt.edu/molbio Retrieve Gene Expression data NCBI GEO EBI ArrayExpress NextBio Research http://www.hsls.pitt.edu/molbio Gene Expression Database http://www.hsls.pitt.edu/molbio Searching GEO http://www.hsls.pitt.edu/molbio Searching GEO http://www.hsls.pitt.edu/molbio Searching GEO http://www.hsls.pitt.edu/molbio GEO2R http://www.hsls.pitt.edu/molbio GEO2R – Experiment to Gene List http://www.hsls.pitt.edu/molbio GEO2R – Experiment to Gene List http://www.hsls.pitt.edu/molbio GEO2R – Experiment to Gene List http://www.hsls.pitt.edu/molbio http://www.hsls.pitt.edu/molbio NextBio Registration https://www.nextbio.com/b/register/register.nb http://www.hsls.pitt.edu/molbio NextBio http://www.hsls.pitt.edu/molbio NextBio http://www.hsls.pitt.edu/molbio NextBio http://www.hsls.pitt.edu/molbio NextBio Search http://www.hsls.pitt.edu/molbio NextBio GeneList http://www.hsls.pitt.edu/molbio Gene Expression Atlas http://www.ebi.ac.uk/gxa/ http://www.hsls.pitt.edu/molbio Array Express http://www.hsls.pitt.edu/guides/genetics Literature to GeneList GEO Geo2R NextBio Gene List http://www.hsls.pitt.edu/molbio Gene Lists to Biology http://www.hsls.pitt.edu/molbio Suggested Reading http://www.ploscollections.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1002820 http://www.hsls.pitt.edu/molbio GO Consortium http://www.hsls.pitt.edu/molbio GO Annotation Cellular Component Biological Process Molecular Function http://www.hsls.pitt.edu/molbio Levels of abstraction Gene Ontology (GO) Khatri, P. et al. Bioinformatics 2005 21:3587-3595; doi:10.1093/bioinformatics/bti565 Copyright restrictions may apply. Evolution history of GO-based functional analysis software Development of gene ontology based tools Khatri, P. et al. Bioinformatics 2005 21:3587-3595; doi:10.1093/bioinformatics/bti565 Copyright restrictions may apply. http://www.hsls.pitt.edu/molbio Gene list to Biology Biological List B-factor, properdin APEX nuclease (multifunctional DNA repair enzyme) 1 ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase) proliferating cell nuclear antigen heterogeneous nuclear ribonucleoprotein A1 fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) three prime repair exonuclease 2 ligase I, DNA, ATP-dependent flap structure-specific endonuclease 1 cyclin A2 cyclin-dependent kinase inhibitor 1A (P21) checkpoint kinase 1 homolog (S. pombe) replication protein A1, 70kDa Werner syndrome exonuclease 1 uracil-DNA glycosylase 2 Functions Pathways Biological Table Name Time 0 Time 15 mins Time 1 hr Time 5 hr IL6 35.45% 36.46% 95.34% 13.91% PDGFRA 32.42% 45.43% 30.99% 56.62% DCN 21.42% 1.29% 33.13% 41.40% ZFP36L1 48.17% 47.56% 84.31% 6.72% DEGS 46.41% 90.30% 52.14% 31.61% GNB2 42.37% 98.25% 25.28% 53.26% ATRN 93.42% 75.90% 1.56% 82.61% PDK4 88.18% 38.03% 95.95% 5.78% MAPK1 20.88% 64.99% 43.18% 52.77% THBS1 99.61% 54.35% 85.28% 85.69% CD68 9.35% 44.81% 69.03% 64.88% DIAPH2 54.16% 34.47% 63.26% 33.46% Sep6 30.12% 48.83% 28.57% 76.28% BFAR 10.18% 7.65% 60.30% 42.66% CCRK 79.03% 19.54% 96.15% 88.79% MT1E 49.07% 23.60% 93.13% 15.83% Interaction Networks http://www.hsls.pitt.edu/molbio Discovery Step: Pathway Analysis Ingenuity IPA DAVID Gene List Metacore http://www.hsls.pitt.edu/molbio Pathway Analysis Software NLP Driven PathwayArchitect PathwayStudio Human Curated Ingenuity IPA GeneGO’s Metacore Biobase Explain “ Axin binds beta-catenin, GSK-3beta and APC.” Extracted Facts: Axin - beta-catenin interaction: Binding Axin - GSK-3beta interaction: Binding Axin - APC interaction: Binding http://www.hsls.pitt.edu/molbio DAVID Bioinformatics Resources http://www.hsls.pitt.edu/molbio DAVID Tools Functional Annotation Clustering Chart – Term centric Table – Gene centric http://www.hsls.pitt.edu/molbio DAVID http://www.nature.com/nprot/journal/v4/n1/fig_tab/nprot.2008.211_T1.html http://www.hsls.pitt.edu/guides/genetics NIH DAVID http://www.hsls.pitt.edu/molbio Ingenuity IPA http://www.hsls.pitt.edu/molbio IPA IPA IPA IPA Function IPA Function IPA Pathways IPA Pathway IPS Upstream Regulator IPA Mechanistic Network IPA Networks IPA Networks IPA GeneGo: Metacore http://www.hsls.pitt.edu/guides/genetics Pathway Analysis: GeneGo Metacore http://www.hsls.pitt.edu/guides/genetics Metacore: Analyze Network (receptors) C-Myc p53 Metacore: Transcription Regulation http://www.hsls.pitt.edu/guides/genetics Cytoscape http://www.hsls.pitt.edu/molbio Hands-on Exercise Use NIH DAVID to uncover enriched pathways associated with the “gene list – 1” http://www.hsls.pitt.edu/molbio Thank you! Any questions? Ansuman Chattopadhyay ansuman@pitt.edu http://www.hsls.pitt.edu/molbio