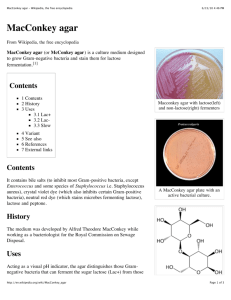
Intestinal Pathogens
advertisement

Name_______________________________ Section______________________________ Introductory Microbiology Pre-Lab Assignment Intestinal Pathogens Note: ALWAYS write scientific names correctly! 1. (1pt) Using your textbook or other resources, describe the difference between commensal and mutualistic organisms. 2. (1pt) How can the fecal-oral cycle of disease be broken? 3. (1pt) List the two genera of enteric pathogens of prime medical concern used in this experiment. 4. (1pt) List two examples given in this lab of coliforms. 5. (1pt) Which of the above (questions 3 & 4) are… a. Non-lactose fermenters: b. Lactose fermenters: 1 10/07/15 2 Lab Exercise: Intestinal Pathogens In this exercise you will examine the morphological and biochemical features of intestinal pathogens. The microbial populations of the large intestine are enormous. It is estimated that there are approximately one TRILLION bacterial cells per gram of feces (that’s about the weight of one paperclip). Most of the intestinal organisms (enterics) are commensals, and many are in mutualistic relationships with their human hosts. Some intestinal bacteria synthesize useful vitamins, such as folic acid and vitamin K. In addition, the normal intestinal microbiota prevents colonization of pathogenic species by producing antimicrobial substances and by competition (mutual exclusion). The enteric pathogens of prime medical concern are the genera Salmonella and Shigella. They cause enteric fevers, food poisoning, and bacillary dysentery. Salmonella typhi, which causes typhoid fever, is by far the most significant pathogen of the Salmonella group. Shigella species are the prime cause of human dysentery. Most diseases of the gastrointestinal system result from the ingestion of food or water contaminated with pathogenic microorganisms. Good sanitation practices, modern methods of sewage treatment, and the disinfection of drinking water help break the fecal-oral cycle of disease. Since a patient’s feces usually contain many genera of bacteria, not just the etiologic agents of disease, it is necessary to use media that are selective and differential to favor the growth of the pathogens. For example, media has been developed to differentiate between lactose fermenting enterics and non-lactose fermenting enterics. The lactose fermenters are called coliforms and with just a few exceptions, such as some strains of Escherichia coli, are generally not pathogenic. The non-lactose fermenting group includes Salmonella and Shigella. You will be given samples of two unknown intestinal organisms. One will be a lactose fermenting coliform (such as Escherichia or Enterobacter), and the other will be a non-lactose fermenter (either Salmonella or Shigella). These organisms will be of less dangerous strains, but their presence will demand caution in handling. Your problem will be to make a genus identification of both. A separation outline is included (next page) listing a series of tests you will perform that differentiate the lactose and non-lactose fermenters and will allow you to determine which organisms in each group you are working with. In actual practice, a fecal sample would initially be placed in an enrichment broth. Enrichment media serves two purposes. It is selective- it inhibits the growth of nonpathogens commonly found in the intestinal tract and favors (or enriches) the growth of pathogenic enterics. Since we are not using stool samples in this exercise, this enrichment procedure is omitted. Instead, you will inoculate the media used in the first test, Kligler’s iron agar, directly from the cultures provided to you. 3 Separation Outline Lactose Lactose + Lactose - Indole + Citrate + Indole - Citrate - Urea + Glucose + Urea Motile H2S+ - Urea+ Indole+/- Citrobacter Escherichia Klebsiella Enterobacter Proteus Glucose - Nonmotile H2S Indole Urea - Urea Indole - Salmonella Shigella Pseudomonas alcaligenes - 4 Day 1: Differential Media, Fermentation, and Production of Identifying Metabolites Many biochemical tests utilize a bacterial species’ ability to synthesize specific enzymes for identification purposes. Since enzymes catalyze chemical reactions, the products of those reactions can sometimes be detected. For example, if a chemical reaction produces acid, a pH indicator in the medium will change color in the presence of that acid. This would indicate that the bacterium carried out that reaction. Since many bacteria have established enzymes, we can identify these bacterial species by testing a collection of reactions that a specific bacterium is known to undergo. The types of chemical reactions tested often include fermentation reactions or reactions requiring or producing specific metabolites. Metabolites are small molecules produced during or are necessary for metabolism. The media used contain the reactants of the reaction in question and often an indicator molecule that detects the products. Materials: per group of 2 students 1 unknown culture on a MacConkey agar plate with a label from 1 to 10 1 unknown culture on a MacConkey agar plate with a label from 11-20 2 Kligler iron agar slants inoculating wire & loop Bunsen burner and striker Gloves, safety glasses 1. MacConkey agar morphology (outward appearance) observations. There are several differential media that have been developed for the isolation of intestinal pathogens. MacConkey agar, observed today, contains lactose and a dye so that if an organism is a lactose fermenter, its colony will take on the reddish color of the dye. Since coliforms ferment lactose, they produce reddish colonies that may take on a mucoid appearance, or dark centered colonies. Non-lactose fermenters, such as Salmonella and Shigella (the socalled SS enterics), produce smooth, colorless colonies that are not reddish/pink. Obtain 1 unknown culture on a MacConkey agar plate with a label from 1 to 10 Obtain another unknown culture on a MacConkey agar plate with a label from 11-20 –Procedure: Observe the morphology of both of your unknowns’ colonies on the MacConkey agar plates. Look for isolated colonies. Determine which plate contains colonies characteristic for Salmonella or Shigella organisms (smooth, colorless colonies) and which plate contains colonies characteristic of coliforms (reddish, mucoid, or dark centered colonies). Using the morphology results of your unknowns, make a preliminary determination of which of your unknowns is a lactose fermenter and which is not. Record your results in the lab report now. Be sure to write your unknowns’ numbers in the lab report results table (#4 in lab report). 5 2. Kligler iron agar test. You will inoculate Kligler iron agar slants for further differentiation. Kligler iron agar contains two sugars, glucose and lactose. Therefore, you will be able to confirm your lactose fermentation results from the MacConkey agar. In addition, iron also is included in order to detect H2S production. A pH indicator in the media will cause it to turn from red to yellow if each sugar is fermented while a black precipitate will indicate if the organism is a producer of H2S. Obtain 2 Kligler iron agar slants. Label the Kligler iron agar tubes with the ‘unknown numbers’ found on the MacConkey agar plates, along with your lab section, your initials, the date, and media type. Do NOT mix up your numbers! -Procedure: With much care and aseptic technique, use a straight inoculating wire to streakstab each Kligler iron agar slant with the appropriate bacterial culture (move the wire over the entire surface of the slant in a zig-zag motion before stabbing). -Incubate the slants at 370C. -Refrigerate the unknown MacConkey agar plates for use on Day 2. 6 Day 2 First, you will determine the fermentation and H2S results of both of your unknowns from the Kligler iron agar slants and compare the Kligler results with your determination from the MacConkey plates. Then you will begin several more tests to finalize the identity (the genus) of your coliform and your non-lactose fermenter. Materials: per group of 2 students: Your MacConkey agar plates from previous lab 2 Kligler iron agar slants inoculated during the previous lab 2 Urea broth tubes 2 SIM tubes 2 Simmons citrate agar slants-Slants should be room temperature at the time students inoculate them. Inoculating wire and loop Bunsen burner, Striker, Gloves, Safety glasses Obtain your Kligler iron agar slants from the previous lab (these may be in the refrigerator instead of the incubator) 1. Kligler iron agar results. Examine the Kligler agar slants from the previous period. You will observe the slant area of the tube and the butt area of the tube. a. Lactose fermentation. The slant area of the Kligler iron agar slant contains lactose. If the slant is yellow, the organism ferments lactose. Sometimes a poor or false lactose fermentation result may occur. If the organism strongly fermented glucose in the butt, some of the resulting acid could leak up into the slant. Compare your lactose results in the Kligler slant to your MacConkey plate results. Do they match? They should. b. Glucose fermentation. The butt of the Kligler iron agar tube contains glucose. Tubes that have a yellow butt ferment glucose. Note: Occasionally a poor glucose result may occur (yellow not present or poorly discernable in butt, even though the organism ferments glucose). All of the organisms used for student unknowns in this lab DO ferment glucose, so a poor result may occur due to low levels of glucose fermentation or coverage of most of the yellow with the production of the black H2S. c. A black precipitate in the medium will indicate that the organism is an H2S producer. d. Use the Interpretation on the page 10 to determine your results. Record all results in the lab report (#4). 7 Obtain 2 of each of the following (one of each for each unknown): Urea broth, SIM media, Simmons citrate slants Label each of the new tubes with your unknowns’ numbers, your lab section, the media type, and your initials 2. Urea broth. Urea broth is a differential medium that tests the ability of an organism to produce the exoenzyme urease that hydrolyzes urea to ammonia and carbon dioxide. The broth contains urea, a small amount of nutrients for the bacteria, and the pH indicator phenol red. Phenol red turns yellow in an acidic environment and fuchsia in an alkaline environment. If the urea in the broth is degraded and ammonia is produced, an alkaline environment is created, and the media turns pink. -Procedure: With a loop, inoculate appropriate urea broth tubes using a colony from each Kligler slant (or MacConkey agar plate, if needed). 3. SIM media. You will inoculate tubes of SIM (sulfur-indole-motility) media to determine 1. motility, 2. hydrogen sulfide production, and 3. indole production. The indole test is a biochemical test performed to determine the ability of the organism to split indole from the amino acid tryptophan. This division is performed by a series of different intracellular enzymes, a system generally referred to as "tryptophanase." -Procedure: With a straight wire, stab a colony from each of the unknowns into appropriate SIM tubes. Stab in the center to 2/3 of the depth of the medium. Pull the straight wire out of the same hole you stabbed it in. 4. Simmons citrate agar slants. Citrate, a Krebs cycle intermediate, is generated by many bacteria; however, utilization of external citrate in the environment requires the presence of citrate transport proteins (permeases). In the citrate test, citrate is the sole source of carbon in the Simmons citrate medium while inorganic ammonium salt (NH4H2PO4) is the sole fixed nitrogen source. When an organic acid such as citrate is used as a carbon and energy source, usually alkaline carbonates and bicarbonates are produced. The visible presence of growth on the medium or visible growth with a change in the pH indicator color due to the increased pH are the signs that an organism can import citrate and use it as a sole carbon and energy source; such organisms are considered to be citrate positive. -Procedure: Slants should be room temperature at the time students inoculate them. With a straight wire, lightly streak each of your unknowns on the surface (from bottom to top with a straight streak) of a different Simmons citrate agar slant. Oxygen is needed for citrate utilization (it is used in the Krebs cycle) so place the caps on these tubes LOOSELY; do not screw them shut tightly. 8 Incubate all tubes at 370C Day 3: Final Evaluation Today, the tubes of SIM medium, urea broth, and Simmons citrate agar slants will be evaluated. You will determine which of your unknowns was a coliform, which was a non-lactose fermenter, and both genus identifications. Materials: per group of 2 students: Tubes of SIM medium, urea broth, and Simmons citrate agar from previous period Kovacs’reagent Use the Interpretation on the following pages to determine your results. Record all results in the lab report (#4). 1. SIM medium. a. First, check for motility. If you see cloudiness spreading away from the stab line, the organism is motile. b. Second, check for H2S production. A black precipitate will be evidence of H2S production. c. Third, test for indole production by adding a few drops of Kovacs’ reagent into the SIM medium. A pink to deep red or violet color in the surface alcohol layer of the broth will develop if indole is produced. A negative result appears yellow. A variable result can also occur, showing an orange color as a result. This is due to the presence of skatole, another possible product of tryptophan degradation. 2. Urea broth. Examine the urea broth tubes. If the medium has changed from yellow to red or deep pink, the organism is urease positive. 3. Simmons citrate slant. If the medium has turned from green to an intense Prussian blue, the organism is citrate. 9 Selective and Differential Test Interpretation MacConkey Agar Colony morphology Result Interpretation Symbol smooth, colorless colonies non-lactose fermenter lactose - reddish, mucoid, or dark centered lactose-fermenter lactose + Kligler’s Iron http://www.bd.com/europe/regulatory/Assets/IFU/Difco_BBL/211317.pdf Result Interpretation Symbol red slant/red butt no fermentation lactose & glucose - red slant/yellow butt glucose fermentation glucose+, lactose- yellow slant/yellow butt glucose and lactose fermentation glucose+, lactose+ black in agar H2S production H2S + SIM Medium http://iws2.collin.edu/dcain/CCCCD%20Micro/sim_deep.htm Sulfur Reduction Results Result Interpretation Symbol black in the medium H2S production + no black in the medium sulfur is not reduced - Motility Results Result Interpretation Symbol growth has spread from stab line motility + little spreading of growth nonmotile 10 Indole Production Results Result Interpretation Symbol red after addition of Kovacs’ reagent tryptophane converted to indole + no color change after Kovacs’ reagent tryptophane not converted to indole - http://en.wikipedia.org/wiki/Indole_test Simmons citrate medium Result Interpretation Symbol growth is visible or growth with blue color blue color organism can import citrate and use it as a sole carbon and energy source + no growth and media remains green organism cannot import citrate and use it as a sole carbon and energy source - http://www.bing.com/images/search?q=simmons+citrate&qpvt=simmons+citrate&FORM=IGRE #view=detail&id=E8229A40467F5A71C11C444A32D33C2C349312EE&selectedIndex=4 To determine the genera of your unknowns, use the separation outline near the beginning of the lab. START with lactose fermentation and work your way down the tests in the outline. 11 12 Name__________________________ Lab Section: Intestinal Pathogens Lab Report 1. (1pt) What is the fermentation characteristic that separates the SS (Salmonella/Shigella) pathogens from the coliforms (such as E. coli and Enterobacter)? SS pathogens:_______________________________________________ Coliforms: 2. (1pt) Describe the expected appearance of each of the following organisms on MacConkey agar. Look in the lab’s description of what the coliform and the SS bacterial morphologies should look like on this medium. Coliforms________________________________________________________ SS pathogens________________________________________________________ 3. (5pt) Using the Separation Outline, fill in the table below with the expected test results (+or-) for these organisms. Expected results not listed in the Separation Outline can be found in Bergey’s Manual of Determinative Bacteriology (Found in the lab- look up the organisms in the index and go to the bolded pages first). Test Salmonella Lactose Glucose Motility H2S production Indole Urea Citrate Shigella Proteus E. coli Enterobacter 13 4. (2pt) Record all of the actual results of your unknowns’ in the table below. Medium MacConkey Agar Unknown #__________ Write the colony morphology: Unknown #__________ Write the colony morphology: Circle one: Is this morphology on MacConkey agar indicative of a lactose fermenter or a non-lactose fermenter? Circle one: Is this morphology on MacConkey agar indicative of a lactose fermenter or a non-lactose fermenter? Lactose fermentation_____ Lactose fermentation_____ Does this result match with the lactose fermentation results from the MacConkey agar? Does this result match with the lactose fermentation results from the MacConkey agar? Glucose fermentation_____ Glucose fermentation_____ H2S _____ Sulfur (H2S)_____ H2S_____ Sulfur (H2S)_____ Motility_____ Motility _____ Indole_____ Indole _____ See your answer to question #1. Kligler Iron agar Indicate + or – SIM MediumIndicate + or - Urea + or Citrate + or - 5. (1pt) What is the genus of each of your two unknowns? Coliform Or Non-lactose fermenter? Unknown # Genus Unknown # Genus 14