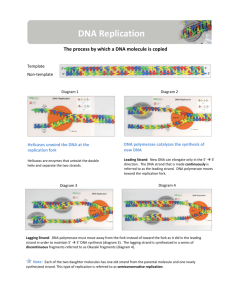
topic 7.2 step diagram
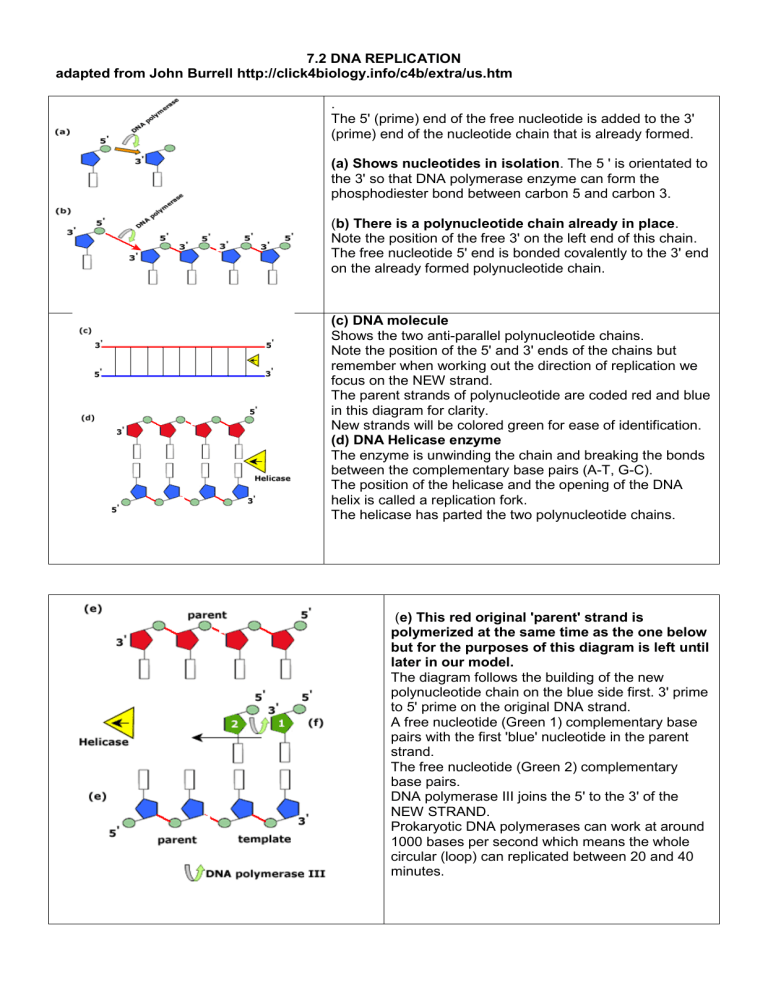
7.2 DNA REPLICATION adapted from John Burrell http://click4biology.info/c4b/extra/us.htm
.
The 5' (prime) end of the free nucleotide is added to the 3'
(prime) end of the nucleotide chain that is already formed.
(a) Shows nucleotides in isolation . The 5 ' is orientated to the 3' so that DNA polymerase enzyme can form the phosphodiester bond between carbon 5 and carbon 3.
( b) There is a polynucleotide chain already in place .
Note the position of the free 3' on the left end of this chain.
The free nucleotide 5' end is bonded covalently to the 3' end on the already formed polynucleotide chain.
(c) DNA molecule
Shows the two anti-parallel polynucleotide chains.
Note the position of the 5' and 3' ends of the chains but remember when working out the direction of replication we focus on the NEW strand.
The parent strands of polynucleotide are coded red and blue in this diagram for clarity.
New strands will be colored green for ease of identification.
(d) DNA Helicase enzyme
The enzyme is unwinding the chain and breaking the bonds between the complementary base pairs (A-T, G-C).
The position of the helicase and the opening of the DNA helix is called a replication fork.
The helicase has parted the two polynucleotide chains.
( e) This red original 'parent' strand is polymerized at the same time as the one below but for the purposes of this diagram is left until later in our model.
The diagram follows the building of the new polynucleotide chain on the blue side first. 3' prime to 5' prime on the original DNA strand.
A free nucleotide (Green 1) complementary base pairs with the first 'blue' nucleotide in the parent strand.
The free nucleotide (Green 2) complementary base pairs.
DNA polymerase III joins the 5' to the 3' of the
NEW STRAND.
Prokaryotic DNA polymerases can work at around
1000 bases per second which means the whole circular (loop) can replicated between 20 and 40 minutes.
( g) The nucleotide sequence is building up as a new polynucleotide using the original as a template.
Specificity is maintained through complementary base pairing of A-T, G-C.
The next nucleotide (Green 3) has base paired and is being polymerized by DNA polymerase III.
The helicase is progressing just ahead of the DNA polymerase III creating the replication fork.
Errors do occur in the replication process but there are biochemical proof reading, repairing and removal mechanisms.
Retuning to the other parent polynucleotide (red).
Since this was anti-parallel to the blue strand the template nucleotides (red) have the opposite direction to the blue ones.
(h) Free nucleotides have complementary base paired with the first two template bases.
Notice that the nucleotides cannot be joined as
DNA polymerase is specific to joining 5' to 3'.
DNA polymerase III has an active site that is specific to the 5' to 3' orientation of the two nucleotides to be joined.
The chain is not polymerized at this stage.
The bases continue to add working in behind the
DNA helicase enzyme. (Upper Green 1-4).
Below we have the polymerization of the new strand on the 'blue' template as previously described in part (g).
(L) The red strand in which the nucleotides do not immediately polymerize is called the
Lagging strand and it forms on the "5 to 3' original DNA template.
(m) Note that there are now a number of bases already in position forming the lagging strand.
DNA polymerase III can now work 'backwards' towards the ori joining the sugar phosphate backbone of the polynucleotide.
On the lagging strand the DNA polymerase is working away from the replication fork.
(n) On the Leading strand the DNA polymerase
III works towards or follows the helicase .
The important point to note here is that the DNA polymerase only works by joining 5' nucleotides to
3' nucleotides on the established chain.
The lagging strand is therefore made up of a number of short polynucleotide chains that need joining together. The short chains are called
Okazaki fragments after the Japanese Biochemist
Reiji Okazaki.
Summary:
This diagram is the usual version used to describe the process of leading and lagging strand polymerase activity.
( p) shows the orientation of the DNA helix
(with helicase) for the diagram below.
(q) Note the position of the replication fork with the DNA helicase opening the DNA chain.
(r) The leading strand forming with DNA polymerase III
On the lagging strand (top green) the new strand is presented as a number of Okazaki fragments.
( s) The DNA polymerase III on this strand has to work from the beginning of each fragment towards the 3' free end of the lagging strand (away from the replication fork).
DNA ligase is the enzyme that joins the fragments.
Remember always think about the action of the
DNA polymerase III adding the 5' of the free nucleotide to the 3' of the already established new strand. This single fact allows the process to be tracked and alternative diagrams to be interpreted.
Primers:
In fact all polymerization both leading and lagging strands actually begin with the addition of 'priming'
RNA nucleotides.
(u) RNA nucleotides (yellow) attach to the first few bases on the template through the action of a
Primase enzyme.
DNA polymerase III then adds DNA nucleotides to the Primer (v).
Later the RNA primer is broken down and removed by DNA polymerase I
DNA nucleotides are added to replace the removed RNA nucleotides.
The Pentose -phosphate backbone is joined by DNA ligase.