Lab: Genetics of Corn - Ms. Dooley's Science Class
advertisement

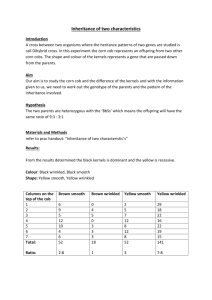
Lab: Genetics of Corn (dihybrid cross) Question: How can the phenotypes of F2 offspring be used to determine the genotypes and phenotypes of the F1 and P generations? Big Idea: The genotypes of the parents and offspring can be determined by looking at the phenotype ratios seen in the actual offspring and using a Punnett square. Procedure: (groups of 2) 1. Assign duties: Partner 1 is the recorder. Partner 2 is the corn counter. 2. Carefully obtain one ear of corn. Do NOT remove any corn kernels! Observe the variety of trait combinations: purple (pigmented), yellow (non-pigmented), smooth (starchy), and wrinkled (sweet). There are four combinations possible. 3. Insert a large pin to mark the first kernel to be counted. 4. Count and tally the phenotype of each kernel on the entire corn cob. (NOTE: This must be completed within one class period!) 5. Record the total of each phenotype, as well as the total kernel count for the cob. Experimental Data: Table 1 Phenotype Combinations PURPLE SMOOTH (starchy) PURPLE WRINKLED (sweet) TOTAL OF ALL KERNELS ON COB = YELLOW SMOOTH (starchy) YELLOW WRINKLED (sweet) Data Analysis: The corn grains of the F2 generation (the actual experimental corn kernels you counted) are a result of a cross between … a homozygous purple/starchy parent and a homozygous yellow/sweet parent (P) Use the symbols: purple=P yellow = p P genotypes: smooth=S wrinkled=s ___ ___ ___ ___ x ___ ___ ___ ___ F1 genotype: ___ ___ ___ ___ Four possible genotype combinations for F1: ___ ___ ___ ___ ___ ___ ___ ___ Table 2: Fill in the four genotype combinations on each side. Complete the Punnett Square for the predicted F2 generation. F1 x F1 ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ F2 Genotypes ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ TOTAL = # ___ ___ ___ ___ ___ ___ ___ ___ ___ 16 F2 Phenotypes # purple smooth (starchy) ___ purple wrinkled (sweet) ___ yellow smooth (starchy) ___ yellow wrinkled (sweet) ___ TOTAL = 16 Comparison of Experimental to Predicted Data: 1.) Write in the “Actual Numbers” in Table 3 (see Table 1 phenotype totals). 2.) Calculate the “Predicted Numbers”: Total Kernels on the Cob ___________ divided by the # of possible F2 genotypes (16). _________÷16 = __________ = # per genotype box Now, multiply the # per genotype box x 9 (for purple smooth) = ______ x 3 (for purple wrinkled) = ______ x 3 (for yellow smooth) = ______ x 1 (for yellow wrinkled) = ______ 3.) Record the predictions below, and compare to the actual numbers counted. Table 3 Phenotype Purple Smooth Actual Data Numbers: Predicted Numbers: Purple Wrinkled Yellow Smooth Yellow Wrinkled Do your Actual data numbers and the Predicted numbers seem close? ________ How could you explain why these numbers aren’t exactly the same? _________ _______________________________________________________________ Explanation: In a dihybrid cross of homozygous parents (P), we see that the heterozygous F1 gametes combine to create 16 possible genotypes resulting in four phenotypes with an approximate 9:3:3:1 ratio in the F2 generation: 9 Purple Smooth : 3 Purple Wrinkled : 3 Yellow Smooth: 1 Yellow Wrinkled Now, answer the Research Question: “How can the phenotypes of F2 offspring be used to determine the genotypes and phenotypes of the F1 and P generations?” _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________