codon
advertisement

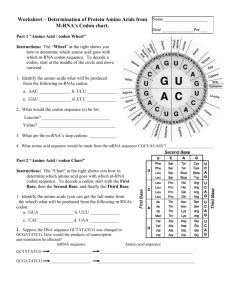
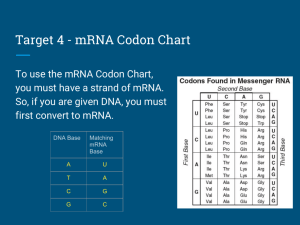
CODON-USAGE A protein sequence can be reverse-translated into i) Fully ambiguous DNA sequence by taking all possible codons for each amino acid; ii) Non-ambiguous DNA sequence by taking a single, most frequently used codon for each amino acid in preferred host cell; iii) Partially ambiguous DNA sequence by taking the rela tively most frequently utilized codons (one or more than one codon for each amino acid) in such a way that these codons can be expressed by a single codon by utilizing ambiguous nucleotides as recommended by NC-IUB (cf. Eur. J. Biochem. 1985, 150:1-5). For searching R. E. sites in protein-coded DNA sequences (nonambiguous or partially ambiguous), user must create a codon usage table, which would be used to generate DNA template from amino acid sequence. For creating codon usage table, user must enter a single codon for each amino acid. The codon may contain ambiguous or non-ambiguous nucleotides. Except the ambiguous amino acids (ie, Arg, Leu, Ser and Terminator) all other amino acids can be represent by single codons. In case of ambiguous amino acids viz., Arg (CGN & AGR), Leu (CTN & TTR), Ser (AGY & TCN) and Ter (TAR & TGA) which cannot be expressed by a single codon, user must select only one of two possible codons. Amino acids and their codons have been shown in Table 1; the Nucleo tides designation is as recommended by NC-IUB (see Table II). The codon usage file Ecoli.cod and Ecoli1.cod which contain respec tively the most frequent codon and relatively more frequent codons for each amino acid, have been included with the program, according to Wada et. al., Nucleic Acids Res. 1992, 20:2111. The user can likewise also create other files with the desired codon usage. Thus a given AA sequence can be optimized with respect to codon usage in the DNA sequence given the preference of different expression systems and/or the secondary structure with mRNA as a result of different codon preferences. ----------------------------------------------------------------Table I. Codon table ----------------------------------------------------------------Amino acid Codon Ambiguous codon ----------------------------------------------------------------Ala Arg Asn Asp Cys Gln Glu Gly His Ile GAC AGA AAC GAC TGC CAA GAA GGA CAC ATA GCC AGG AAT GAT TGT CAG GAG GGC CAT ATC GCG CCT CGA CGC CGG CGT GGG GGT ATT GCN AGR and CGN AAY GAY TGY CAR GAR GGN CAY ATH Leu CTA CTC CTG CTT TTA TTG CTN and TTR •37 •3 ŠLys AAA AAG AAR Met ATG ATG Phe TTC TTT TTY Pro CCA CCC CCG CCT CCN Ser AGC AGT TCA TCC TCG TCT AGY and TCN Thr ACA ACC ACG ACT ACN Trp TGG TGG Tyr TAC TAT TAY Val GTA GTC GTG GTT GTN Terminator TAA TAG TGA TAR and TGA --------------------------------------------------------------------------------------------------------------------------------Table II. Nucleotide designations recommended by the Internaional Union of Biochemistry ----------------------------------------------------------------A = adenosine W = A or T H = A, C, or T G = guanine S = C or G B = G, C, or T C = cytosine K = T or G N = A, G, C or T T = thymidine M = C or A Y = C or T D = A, G, or T R = A or G V = A, G, or C -----------------------------------------------------------------