Physiological Characteristics of the Unknown
advertisement

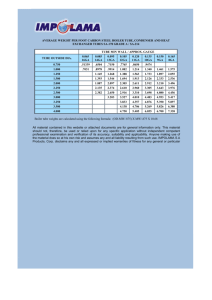
Name: Section: Unknown #: Introductory Microbiology Prelab Assignment Biochemical (Physiological) Characteristics of the Unknown 1. (0.5pt.) What is the difference between selective and differential media? 2. (1pt.) Define ‘positive control’? Why will a positive control be included in the experiments? 3. (0.5pt.) What is the function of a pH indicator? 4. (0.5pt.) Based on the results of the morphological tests, list ALL of the biochemical tests you and your partner will perform on your unknown in this lab (see tables on p. 5). 1 5. (0.5pt.) Describe a positive result for the lactose fermentation test. 6. (1pt.) List the enzymes tested for in each of the following. Mixed acid fermentation: Indole test: Urea hydrolysis: 7. (1pt.) Describe the difference between a morphological and a biochemical trait. 2 Lab Exercise Biochemical (Physiological) Testing of the Unknown 01/13/16 Biochemical testing Biochemical or physiological tests determine metabolic properties. These include the ability to use specific nutrients, products released during growth, and processes for obtaining energy. These functions are performed through chemical reactions. During a chemical reaction, one or more starting materials, called the reactant(s) are converted to one or more different chemicals, the products. In order for metabolic processes to occur in a time-frame compatible with life, enzyme catalysts speed up the rate of chemical reactions. This occurs when an enzyme’s active site bonds to the reactant(s) which are also called the enzyme’s substrates. Precise substrate positioning on the active site reduces the energy limitations of the reaction so it proceeds at a faster rate. What determines which enzymes a bacterium produces? Genes! Therefore, during the biochemical testing of the unknown, you are indirectly compiling a genetic profile. This is instructive in identifying your unknown because each species has a unique set of genes. Media, reagents, and indicators In order to test for specific chemical reactions (and, therefore, the presence of particular enzymes), the unknown will be inoculated in a variety of media. Selective media favor the growth of certain bacteria but inhibit others. Differential media result in distinctive characteristics based on the particular types of bacteria (such as colony or media color). One can determine whether a microbe synthesizes certain enzymes by detecting the end-products of the reactions they catalyze. Substrates may be included in the media or reagents1 added after incubation. A pH indicator in the medium develops distinctive colors when a change in pH accompanies the reaction. A common pH indicator is phenol red (red under basic conditions, yellow under acidic conditions). (starting substrate) fermentation reaction pathway lactose (end- product) acids (medium changes color) Although very common, not all reactions produce a change in the pH, and other types of indicators are required. For example, alcohol also is an end-product of some types of fermentation. Other reagents produce a distinctive color in the presence of alcohol, thus revealing the results. Positive controls It is very important that the tests include positive controls, organisms that have known characteristics or undergo the reaction in question. Positive controls allow us to observe expected results and serve as media, reagent, and procedural checks. If a positive control yields an unanticipated result, your unknown’s result may also be atypical. The instructor or lab technician will inoculate the positive controls. They are generally found on the lab bench at the back of the room. 1 (substances which allow detection of chemical reactions or serve as reactants) 3 Examples of biochemical tests Many tests detect the presence of enzymes in order to evaluate nutritional and metabolic activities. Specific catabolic processes, used to transform nutrient energy into ATP molecules, can be very informative. The three major catabolic processes are aerobic respiration, anaerobic respiration, and fermentation. All three initially undergo a metabolic pathway called glycolysis where sugars are broken down into pyruvic acid. Respiration can be aerobic or anaerobic, but always incorporates a Krebs cycle and electron transport system. Organisms limited to aerobic respiration for energy are ‘strict aerobes’. On the other hand, some microbes are facultative (often called ‘facultative anaerobes’), meaning they are able to undergo aerobic respiration and anaerobic processes to synthesize ATP. Organisms that cannot undergo aerobic respiration and only utilize anaerobic processes, such as anaerobic respiration and/or fermentations are ‘strict anaerobes’. Most strict anaerobes cannot tolerate the toxic by-products and, therefore, the presence, of O2. Fermentations are the most common catabolic processes used to identify microbes. Fermentative pathways are always anaerobic but never incorporate a Krebs cycle or electron transport chain. Therefore, anaerobic respiration is different from fermentation in the strictest sense. Carbohydrate fermentation media incorporate different sugars to test for the bacterium’s ability to use them as reactants for fermentation. If fermented, the acid/alcohol endproducts can be detected. Other metabolic processes also are determined in biochemical studies, such as the ability to digest substances in hydrolysis reactions (where molecules are broken down in the presence of water), or miscellaneous characteristics indicative of certain species. As a whole, microorganisms have a myriad of extremely diverse metabolic activities. Some are very fastidious (particular) while others may use several approaches. This depends on the genome of the microbe, available nutrients, and environmental conditions. Analyzing results It must be emphasized that we are working with LIVING ORGANISMS. All studies on living organisms have certain pitfalls. One of these complications, especially in bacteria, is strain variation produced through mutation. All strains of one species do not necessarily obtain the same results for every test. Your unknown may not possess exactly the same characteristics as the stock culture from which it was subcultured or the generalized characteristics listed in reference manuals. In addition, the results of some tests will be inconclusive: the positive control results might be atypical, the results may be weak, the media may be unsuitable, procedures were not followed, etc. Do not become frustrated if a few inconclusive results are obtained. You will use the totality of the results to identify the unknown. Determining the scientific name of your unknown Continue using online sites, Bergey's Manual of Systematic Bacteriology, the Difco manual, and other textbooks and resources to document known characteristics of the microbes listed in the Unknown Characteristic Chart. Please do not write in any books in the library or in the lab. After completing the biochemical tests in this lab, record your results in the lab report and the Unknown Characteristics Chart. Remember, your results may not be identical to the characteristics found in manuals or textbooks. Often, manuals or textbooks will list characteristics as occurring "usually", "generally", etc. Continue to narrow down your choices. Eliminate those species most different from yours. 4 First, locate the tables pertaining to your unknown. There are four tables: 1) All Unknowns, 2) Gram Negative Rods, 3) Gram Positive Rods, and 4) Gram Positive Cocci. Everyone will perform the tests in the All Unknowns table. Students must then find the table listing the Gram stain and the shape of their particular unknown. These tables list the specific biochemical tests to be performed by the different types of bacteria. Procedures for each test are found on succeeding pages. All Unknowns All of these tests will be performed on Day 1 Lactose fermentation in lactose broth, p. 6 Sucrose fermentation in sucrose broth, p. 6 Glucose fermentation in glucose broth, p. 6 Mixed acid fermentation test in MR-VP media (methyl red test), p. 7 Voges-Proskauer test in MR-VP media, p. 7 Gram Negative Rods Indole Test: tryptophan hydrolysis in tryptone broth, p. 8 Urea hydrolysis in urea broth, p. 8 Colony color on EMB agar, p. 9 Pigment production on Pseudomonas Isolation Agar, p. 9 Gram Positive Rods Indole Test: tryptophan hydrolysis in tryptone broth, p. 8 Urea hydrolysis in urea broth, p. 8 Catalase test on agar slants, p. 10 Gram Positive Cocci Hemolysis on blood agar, p. 9 Catalase test on agar slants, p. 10 Growth on mannitol salt agar, p. 10 Coagulase test, p. 11 REMEMBER!!!! USE YOUR WORKING STOCK FOR TESTS AND INOCULATIONS, NOT THE RESERVE STOCK! 5 Do not ask the instructor to interpret results until AFTER you compare your unknown’s results to the POSTIVIE CONTROL on the back bench. LACTOSE FERMENTATION TEST: All Unknowns Materials: 1 tube of lactose broth with inverted Durham tube, inoculating loop This test will determine if your unknown contains enzymes catalyzing lactose fermentation. The broth includes a pH indicator. If fermentation occurs, and the media becomes acidic, the broth will turn yellow. The tube also contains a small, inverted Durham tube. Gas bubbles trapped in the Durham tube are another indication of fermentation. Procedure: Label the tube with "lactose", your initials, lab section, and unknown number. Using aseptic technique, inoculate the tube with your unknown. Incubate for 48 hours. Positive result: yellow color (possibly gas in the inverted tube) Positive control: Escherichia coli Negative result: no color change, no gas SUCROSE FERMENTATION TEST: All Unknowns Materials: 1 tube of sucrose broth with inverted Durham tube, inoculating loop This test will determine if your unknown contains enzymes catalyzing sucrose fermentation. The broth includes a pH indicator. If fermentation occurs, and the media becomes acidic, the broth will turn yellow. The tube also contains a small, inverted Durham tube. Gas bubbles in the Durham tube are another indication of fermentation. Procedure: Label the tube with "sucrose", your initials, lab section, and unknown number. Using aseptic technique, inoculate the tube with your unknown. Incubate for 48 hours. Positive result: yellow color (possibly gas in the inverted tube) Positive control: Escherichia coli Negative result: no color change, no gas GLUCOSE FERMENTATION TEST: All Unknowns Materials: 1 tube of glucose broth with inverted Durham tube, inoculating loop This test will determine if your unknown contains enzymes catalyzing glucose fermentation. The broth includes a pH indicator. If fermentation occurs, and the media becomes acidic, the broth will turn yellow. The tube also contains a small, inverted Durham tube. Gas bubbles in the Durham tube are another indication of fermentation. Procedure: Label the tube with "glucose", your initials, lab section, and unknown number. Using aseptic technique, inoculate the tube with your unknown. Incubate for 48 hours. Positive result: yellow color (possibly gas in the inverted tube) Positive control: Escherichia coli Negative result: no color change, no gas 6 MIXED ACID FERMENTATION IN MR-VP MEDIA: (METHYL RED TEST): All Unknowns Materials: 1 tube of MR-VP media, inoculating loop, methyl red indicator Bacteria that ferment glucose in MR-VP media may produce large amounts of different kinds of acids (lactic, acetic, succinic, formic, etc.). This results in a low pH below 5. In addition, the enzyme formic hydrogenylase induces gas formation. Procedure: Label the tube with MR, your initials, lab section number, and unknown number. Inoculate the tube of MR-VP media with your unknown. After 2 – 5 days of incubation, add 5 drops of methyl red indicator Positive result: red color (reaction should occur within seconds) Positive control: Staphylococcus aureus Negative result: yellow color VOGES-PROSKAUER TEST IN MR-VP MEDIA: All Unknowns Materials: 1 tube of MR-VP media, inoculating loop, reagent A, reagent B The Voges-Proskauer test is also known as the butanediol (an alcohol) fermentation test because some bacteria that do not produce acids in MR-VP media instead produce alcohols after fermentation. After incubation, two reagents are added which detect the precursor to butanediol. Procedure: Label the tube of MR-VP media with VP, your initials, lab section, and unknown number. Heavily inoculate the tube with your unknown. After 24 -48 hours of incubation, transfer 1 ml of the MR-VP medium culture to a clean test tube. Add 15 drops of Reagent A (alpha napthol) followed by 5 drops of Reagent B (potassium hydroxide). Shake tube gently, then allow to stand for 20 minutes. Do not read if allowed to stand more than 1 hour.” Positive result: pink or red color Positive control: Enterobacter aerogenes Negative result: no change or a brown color 7 TRYPTOPHAN HYDROLYSIS IN TRYPTONE BROTH (INDOLE TEST): Gram negative rods and Gram positive rods only Materials: 1 tube of tryptone broth, inoculating loop, Kovacs reagent Certain bacteria have the capacity to cleave tryptophan, an amino acid, into indole (a peptone: partially hydrolyzed protein) and pyruvic acid. The enzyme catalyzing this reaction is tryptophanase. The addition of Kovacs reagent will detect indole in the medium. Procedure: Label the tube with indole, your initials, lab section, and unknown number. Inoculate your unknown in one tube of tryptone broth. Incubate for 48 hours. After incubation, add 10-12 drops of Kovacs reagent to the culture. Positive result: red layer formed at the top of the broth Positive control: Proteus vulgaris Negative result: no red layer at the top of the broth UREA HYDROLYSIS IN UREA BROTH: Gram negative rods and Gram positive rods only Materials: 1 tube of urea broth, inoculating loop The enzyme urease cleaves ammonia from the molecule urea. The released ammonia increases the pH causing the phenol red indicator in the medium to change from yellow to red/pink. Procedure: Label the tube with urea, your initials, lab section, and unknown number. Inoculate your unknown in the tube of urea broth. Incubate for 48 hours. Positive result: red/pink color Positive control: Proteus vulgaris Negative result: no color change 8 HEMOLYSIS ON BLOOD AGAR: Gram positive cocci only Materials: 1 plate of blood agar, inoculating loop The lysis of red blood cells by secreted enzymes is called hemolysis. Some species completely lyse red blood cells. This action is called beta hemolysis and results in clear zones around colonies grown on blood agar. Other species have enzymes that transform hemoglobin to a chemical that has a greenish-brown color. This is called alpha hemolysis. Finally, some species do not react with red blood cells at all (no change, also called gamma hemolysis). Procedure: Label your plate with hemolysis, your initials, lab section, and unknown number. Quadrant streak your unknown on a plate of blood agar. Because some species produce hemolysins that are only active in conditions of low oxygen, obtain more bacteria on your loop and ‘stab’ the agar 5 times, apart from the streak. Stabbing creates a ‘pocket’ of low oxygen. Incubate the plate for 48 hours. After incubation, observe the plates for hemolysis. If hemolysis occurred, determine whether it is alpha or beta. Examples of beta and alpha hemolysis will be available for comparison. COLONY COLOR ON LEVINE’S EMB AGAR: Gram negative rods only Materials: 1 EMB agar plate, inoculating loop Levine’s EMB (eosin methylene blue) agar is both selective and differential. It is useful in the identification of enteric (intestinal) coliform bacteria because it inhibits the growth of Gram positive bacteria and causes coliforms to express distinctive colony characteristics. E. coli produces fluorescent green colored colonies due to vigorous fermentation of glucose and lactose in the presence of specific pH indicators. Enterobacter usually exhibits large colonies with dark centers. Procedure: Label the plate with EMB, your initials, lab section, and unknown number. Quadrant streak your unknown on a plate of EMB agar. Incubate at 48 hours. After incubation, observe the color of the colonies. Distinctive plates will available in the lab for observation. See the Difco Manual for colony description. PIGMENT PRODUCTION ON PSEUDOMONAS ISOLATION AGAR: Gram negative rods only Materials: 1 plate of Pseudomonas isolation agar, inoculating loop Pseudomonas isolation agar contains irgasan, a broad-spectrum antibiotic not active against Pseudomonas. In addition, the medium is formulated to enhance the formation of a blue or blue-green pigment by Pseudomonas, which diffuses into the surrounding agar. Procedure: Label the plate with Pseudomonas, your initials, lab section, and unknown number. Quadrant streak the plate with your unknown. Incubate for 48 hours. 9 Positive control: Pseudomonas aeruginosa CATALASE TEST ON TRYPTICASE SOY AGAR SLANTS: Gram positive cocci and Gram positive rods Materials: 1 trypticase soy agar slant, inoculating loop, hydrogen peroxide In order for strict aerobes or facultative anaerobes to undergo respiration, O 2 must be utilized. However, in the cell, oxygen is converted into toxic hydrogen peroxide (H2O2) that must be broken down into nontoxic forms. In order to accomplish this, the enzyme catalase is employed to catalyze the following reaction: Catalase 2H2O2 2H2O + O2 Procedure: Label the tube with catalase, your initials, lab section, and unknown number. With an inoculating loop, zigzag streak the agar slant with your unknown. Use a heavy inoculum. Incubate for 48 hours. After 48 hours, place a few drops of hydrogen peroxide on your colonies. Positive result: bubbling and fizzing resulting from the breakdown of H2O2 Positive control: Micrococcus luteus Negative result: no bubbling or fizzing GROWTH ON MANNITOL SALT AGAR (MSA): Gram positive cocci only Materials: 1 plate of mannitol salt agar, inoculating loop Mannitol salt agar contains high concentrations of sodium chloride. Therefore, it is selective for bacteria that can withstand high salt concentrations. In addition, the presence of mannitol allows for fermentation of this sugar. The pH indicator, phenol red, changes color from red to yellow (acidic conditions) if fermentation occurs. Procedure: Label the plate with mannitol, your initials, lab section, and unknown number. Quadrant streak the mannitol salt agar with your unknown. Incubate for 48 hours. Results: Record both of the following. A. Observe the plates for growth: -growth=tolerance for high salt concentrations -no growth=low tolerance for high salt concentrations B. If growth occurred, observe the medium for mannitol fermentation: -yellow=fermentation occurred -red=no fermentation Positive control: Staphylococcus aureus 10 See the Difco Manual for colony descriptions. 11 COAGULASE TEST: Gram positive cocci only Materials: blood plasma, small test tube, 1.0ml pipet, pipet pump, inoculating loop Coagulase, or clumping factor, is an enzyme capable of clotting blood plasma. It converts fibrinogen in plasma into fibrin resulting in coagulation. These clots may be seen with the naked eye. The coagulase test is useful for identifying certain species of Staphylococci. Staphylococcus aureus, the positive control, usually produces coagulase while other Staphylococcus species do not. Procedure: Label the empty test tube with coagulase, your initials, lab section, and unknown number. Pipet 0.5ml of blood plasma into the test tube. HEAVILY inoculate the plasma with your unknown. Incubate at 37C. Check every 30 minutes for agglutination (clumping) by tilting the tube and observing for solidification. You may have to incubate for 48 hours. 11 12 Name: Section: Unknown #: Introductory Microbiology Lab Report Biochemical (Physiological) Characteristics of the Unknown 1. List your unknown’s physiological test results. 2. (0.5pt.) Reactions in which compounds are broken down into smaller units in the presence of water are called reactions. 3. (0.5pt.) List two resources that students may utilize in order to determine the identity of your unknown. 4. (0.5pt.) List one problem that may be encountered when determining the scientific name of your unknown. 5. (0.5pt.) What are two types of molecules that often produced as end-products of fermentation? 13 6. (0.5pt.) How does a facultative anaerobe obtain energy? 7. (1pt.) Is EMB agar selective or differential? Explain your answer. 8. (0.5pt.) Write the change in pH (increase or decrease) that accompanies a positive result for the following tests. Urea hydrolysis: Methyl red test: 9. (1pt.) List the positive controls for the following tests. Mixed acid fermentation: Tryptophan hydrolysis: Urea hydrolysis: Coagulase test: 14