Vector Quantization (VQ)
advertisement

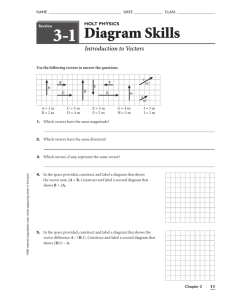
Vector Quantization (VQ) The idea of scalar quantization generalizes immediately to vector quantization (VQ). In this case, we have to perform quantization over blocks of data, instead of a single scalar value. The quantization output is an index value which indicates another data block (vector) from a finite set of vectors, called the codebook. The selected vector is usually an approximation of the input data block. The important point about VQ is that, we require reproduction vectors (instead of reproduction levels) that are known by the encpder and the decoder. The encoder takes an input vector, determines the best representing reproduction vector, and transmits the index of that vector. The decoder takes that index, and forms the reproduction vector because it already knows the reproduction vectors instead of the original. Consider the following figure: The three 2x2 data vectors at the left ( , , ) are quantized to the 2x2 data vector at the right ( ). This means that the encoder transmits the symbol which represents vector when it encounters the 2x2 vectors at the left as its input. Obviously, should be a good representation for the left vectors. The decoder, therefore, reproduces at the places of the original 2x2 vectors at the left. The issues of "how good a representation of the right vector is for the left vectors" is still valid, and the distortion measurements will be similar to the scalar case. The overall encoder and decoder can be given as: The encoder, therefore, just finds the vector in the set from to which is closest to the input vector . Let's say that the closest vector is . The encoder, then transmits the index corresponding to , which is i. The task of the decoder is even easier. It just gets the index i, and extracts the vector from the codebook, which is the same as the codebook of the encoder. The quantized version of is, therefore, . The closest vector to the input vector is found by the nearest neighbor rule. The nearest neighbor encoder selects vector if We use the usual distortion measure, which is the mean squared error (MSE): where, the norm is defined for a vector as: . Exercise (Matlab): Assume that you have the following codebook with three reproduction vectors: -) Generate 8 data vectors (random, if you wish) of size 3x3 all of whose values are between 0 and 1. -) Find the quantization index corresponding to each of your generated data vectors. -) Calculate the amount of reproduction error for each of your data vector. Can you obtain a lower reproduction error if you use different quantization indexes with the same codebook? Why? The adventage of VQ over SQ: In general, the VQ scheme outperforms scalar quantization (SQ). This is reasonable, because SQ is actually a special case of VQ, with vector size=1. Consider a 64x64 256-level (8bits/pixel) gray-scale image (which is a 64x64 matrix). If we do scalar quantization with 8 reproduction levels, the amount of bits required to represent each pixel becomes . This corresponds to a compression of 3:8. On the other hand, if we make a VQ of 2x2 data blocks, and if our codebook has 8 vectors, then each vector can be represented with 3 bits. But how many 2x2 blocks are there in the 64x64 image? Answer is 32x32. So, the quantized data has 32x32x3 bits in total, whereas the original data had 64x64x8 bits. So, the compression ratio is 3:2x2x8 = 3:32, which is much better compression! But are the qualities (or distortions) of the above schemes the same? Usually not. Actually, if the four elements of a vector in the above example are uncorrelated, we would need 4 times the number of codebook vectors used in the scalar quantizer to achieve the same distortion: codebook size of the VQ should be 8x4=32. As a result, the compression ratio would be the same. Fortunately, most of the data have some sort of correlation among its samples. Consider a data composed of the weight and height of a person. The data have the following distribution: Case 1: Separate quantization of the weight and height using uniform scalar quantizers: Since the height data is uniform between 1.60m and 1.80m, and the weight data is uniform between 60kg and 100kg, the 8 level uniform quantizers are: We use 3 bits for representing the height and 3 bits for representing the weight. To represent both, we need 6 bits. Case 2: Vector quantization of pairs . First consider the 64 region uniform quantizer (this means, we use 6 bits to represent the pair): This really corresponds to the scalar quantization of each data (height and weight) separately. As you see, some of the quantization regions correspond to no data. We do not have to use those regions in our codebook to represent anything. If we get rid of the empty regions, we come up with the following codebook: which has only 30 vectors, so 5 bits is enough to represent each codebook vector instead of 6. We obviously have an improvement in efficiency. The efficiency would not improve if the data was scattered uniformly over the weight-height distribution (meaning no cross correlation). Optimum VQ design : Optimality constraints set by Lloyd immediately applies to the vector quantizatin case. As a result, all the equations about the optimal scalar quantizers are valid vor the VQs. Similarly, the Lloyd-Max iterative algorithm is also used for designing optimal VQs. Assume you have N vectors to quantize to M reconstruction vectors, so the codebook size will be M. 1. Start with an initial set of reconstruction vectors and set k=0, 2. The reconstruction vectors define quantization regions with the rule: Eq. V1 This rule corresponds to selecting regions nearest to the reconstruction vectors, also called the Voronoi Cells. A property of the Voronoi cells is that, they are convex regions. An example Voronoi cells illustration for the two element vector case is given as: : Here, dots represent reproduction vectors and the lines are the half ways between each dot. You can see that each region is a convex one, meaning that a line connecting every two points inside the region remains inside the region. The equation for convexity is: given two points x and y inside the convex region, a point also remains in the region for all between 0 and 1. (Exercise: show that the regions described by Equation (Eq. V1) are convex, using the above definition.) 3. Compute the average distortion between the reconstruction vectors and the input data. 4. If then stop, otherwise continue. 5. Increment k. Find new reconstruction vectors vectors inside each quantization region by averaging the input and go to step 2. When the algorithm stops, the Voronoi cells illustration given above has the following properties: The dots (reproduction vectors) are at the centroids (or the averages) of the input data which fall inside the corresponding Voronoi region. The lines are half ways between each black dot. It gets more and more difficult if you proceed to higher dimensions (3 or more data in a vector). For 3D case, the voronoi cells become 3D convex volumes, for more dimensional data, they are convex hyper-regions. Finally, you can download the following Matlab functions which perform optimal 2D vector quantization on images. The programs are: vq.m centroid.m nearest.m The vq.m function is the main function which asks for the input image, the vector size N for extracting NxN portions of the input image (as an example, you give the value 4, and the VQ uses vectors of size 4x4), and the maximum number of vectors to exist in the codebook (which determines the compression ratio). The other functions (centroid and nearest) are called from vq.m to perform the associated operation in the Lloyd-Max quantizer. Please carefully examine the programs, their usage, and the algorithm inside. You can use the lena_small image to experiment on with these programs. I strongly recommend that you also generate the following data: where and the random variables are , , and using the following Matlab commands : . theta=rand(1,200); er1=(rand(1,200)-0.5)/5; er2=(rand(1,200)-0.5)/5; x=[cos(2*pi*theta)+er1;sin(2*pi*theta)+er2]; x1=x(1,:); x2=x(2,:); plot(x1,x2,'r.'); and vector quantize it using 2-element vectors. Before starting quantization, plot the data by saying: x1=x(1,:); x2=x(2,:); plot(x1,x2,'r.'); You should obtain a ring shaped figure: After vector quantization, your codebook entries, ideally, should lie on this ring with almost equal separations. Plot your VQ outputs also, and see if the above argument is correct. If you do not obtain something like that, change your initial vectors and adjust your stopping criterion to achieve something similar to the ideal case. There are some other quantizer design techniques that are developed for specific situations. Some of these techniques are covered inside the common quantizer techniques section.