Alignment with Non-Overlapping Inversions on Two Strings
advertisement

Alignment with Non-Overlapping Inversions on
Two Strings
Da-Jung Cho, Yo-Sub Han, and Hwee Kim
Department of Computer Science, Yonsei University
50, Yonsei-Ro, Seodaemun-Gu, Seoul 120-749, Korea
{dajung,emmous,kimhwee}@cs.yonsei.ac.kr
Abstract. The inversion is one of the important operations in bio sequence analysis and the sequence alignment problem is well studied for
bio sequence comparisons. This gives rise to the alignment with nonoverlapping inversion problem: Given two strings x and y, does there
exist an alignment with non-overlapping inversions for x and y. We, in
particular, consider the alignment problem when non-overlapping inversion are allowed for both x and y. Based on the properties of the inversion
operation, we design an O(n3 ) algorithm using O(n2 ) space, where n is
the size of the input string.
1
Introduction
In modern biology, determining the exact order of DNA sequence and retrieving information of DNA sequence are important problems and alignments of
sequences are widely used for biological sequence comparisons [1, 7, 12, 13]. For
a DNA sequence, a chromosomal translocation is to relocate a piece of the DNA
sequence from one place to another and, thus, rearrange the DNA sequence [9].
The chromosomal translocation is a crucial operation in DNA since it alters
a DNA sequence and often causes genetic diseases [10]. A chromosomal inversion occurs when a single chromosome undergoes breakage and rearrangement
within itself [11]. Based on the important biological events such as translocation and inversion, there is a well-defined string matching problem: given two
strings and translocation or inversion, the string matching problem is finding all
matched strings allowing translocations or inversions. Moreover, people proposed
an alignment with translocation or inversion problem, which is closely related to
find similarity between two given strings; that is to obtain minimal occurrences
of translocation or inversion that transform one to another, applying to one
string. Many researchers investigated efficient algorithms for this problem [1–4,
6, 8, 13, 14].
The inversions, which are one of the important biological operations, are not
automatically detected by the traditional alignment algorithms [14]. Schöniger
and Waterman [13] introduced the alignment problem with non-overlapping inversions, defined a simplification hypothesis that all regions will not be allowed
to overlap and showed an O(n6 ) time algorithm that computes local alignments
with inversions between two strings of length n and m based on the dynamic programming. Vellozo et al. [14] presented an O(n2 m) algorithm, which improved
the previous algorithm by Schöniger and Waterman. They built a matrix for
one string and partially inverted string using table filling method with regard
to the extended edit graph. Recently, Cantone et al. [1] introduced O(nm) time
algorithm using O(m2 ) space for string matching problem, which is to find all
locations in a given text of length n at which a given pattern of length m matches
with non-overlapping inversions.
Many diseases are often caused by genetic mutations, which can be inherited
through generations and can result in new sequences from a normal gene [5]. In
other words, we can have two different sequences from a normal gene by different
mutations. This motivates us to examine the problem of deciding whether or not
two gene sequences are mutated from the same gene sequence. In particular, we
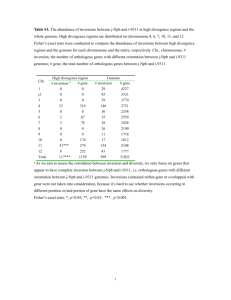
consider an inversion mutation. See Fig. 1 for an example.
T
G C G A G G G T
x(1,2)
Θ(x) =
x(5,7)
C G G C G T
G A
···
x
C T
A C G
Θ(x) = Θ(y)
y(n−3,n−1)
y(7,8)
C A A
A C A C
x(n−1,n)
x(9,9)
C A C G C C T
y(2,6)
···
···
C G T
T
y
Fig. 1. An example of non-overlapping inversions on both strings x and y, where Θx =
(1, 2)(3, 3)′ (4, 4)′ (5, 7)(8, 8)′ (9, 9) · · · (n − 1, n) and Θy = (1, 1)′ (2, 6)(7, 8)(9, 9)′ · · · (n −
3, n − 1)(n, n)′ . Note that (i, i)′ denotes the alignment at position i without complementing x[i].
Note that this problem is different from the previous approaches [13, 14],
where a non-overlapping inversion occurs in one string and transforms it to the
other string; namely Θ(x) = y for a set Θ of non-overlapping inversions. On the
other hand, we consider more general case where inversions can occur in both x
and y simultaneously.
2
Preliminaries
Let A[a1 ][a2 ] · · · [an ] be an n-dimensional array, where the size of each dimension is ai for 1 ≤ i ≤ n. Let A[i1 ][i2 ] · · · [in ] be the element of A with indices
(i1 , i2 , . . . , in ). Given a finite set Σ of character and a string s over Σ, we use |s|
to denote the length of s and s[i] to denote the symbol of s at position i. We use
s(i,j) to denote a substring s[i]s[i + 1] · · · s[j], where 1 ≤ i ≤ j ≤ |s|. We consider
biological operation inversion θ and denote by θ(s) the reverse and complement
of given string s. For example, θ(A) = T and θ(AGG) = θ(G)θ(G)θ(A) = CCT .
We define an inversion operation θ(i,j) for a given range i, j as follows:
θ(i,j) (s) = θ(s(i,j) ).
For simplicity, we use (i, j) instead of θ(i,j) if the notation is clear in the
context. We say that i+j
2 is the center of the inversion for (i, j). We define a
set Θ of non-overlapping inversion to be
Θ = {(i, j) | 1 ≤ i ≤ j and for ∀(i′ , j ′ ) ̸= (i, j) ∈ Θ, j < i′ or j ′ < i}.
Then, for a set Θ of non-overlapping inversions and a string s, we have Θ(s) = s′
where
{
θ(s[j + k − i]) if (j, k) ∈ Θ and j ≤ i ≤ k
′
s [i] =
s[i]
otherwise.
For example, if Θ = {(1, 1), (2, 3)} and s = AGCC, then Θ(s) = θ(A)θ(GC)C =
T GCC. From now on, we use a set of inversions instead of a set of non-overlapping
inversions since we only consider sets of non-overlapping inversions.
Definition 1 (Problem). We formally define a new alignment problem with
non-overlapping inversions on two strings as follows: Given two strings x and y
of the same length, can we determine whether or not there exist two sets Θx and
Θy of inversions such that Θx (x) = Θy (y)?
3
The Algorithm
Vellozo et al. [14] considered the problem of finding an alignment of non-overlapping
inversions on only one string, and proposed an O(n2 m) algorithm using O(n2 )
space for given two strings s and t of length n and m, respectively. We examine
the problem in which non-overlapping inversions are allowed for both strings
of the same length. Note that the problem is more difficult than the problem
considered by Vellozo et al. [14], since we can apply two sets of non-overlapping
inversions Θx and Θy to x and y, respectively.
We use x = AGCT and y = CGAA as our example strings for explaining
the algorithm. Remark that θ(AG)Cθ(T ) = CT CA = Cθ(GA)A and, thus, we
have two sets Θx = {(1, 2), (4, 4)} and Θy = {(2, 3)}.
We start from building a table in which each cell contains a pair of a range
and a character in each index of an input string. We define an array Tx [n][n + 1]
for x as follows:
if j < i,
((j, i), θ(x[j]))
Tx [i][j] = ((i, i)′ , x[i])
if j = i,
((i, j − 1), θ(x[j − 1])) if j > i.
We call all elements in Tx inversion fragments of x. For an inversion fragment F = ((p, q), σ) or ((p, p)′ , σ), we say that F yields the character σ. Inversion
fragments become useful to compute a substring created by any inversion because of the following property of the inversion operation:
θ(i−1,j+1) (x) = θ(x[j + 1])θ(i,j) (x)θ(x[i − 1]).
Observation 1 For a string x and its Tx ,
1. θ(i,j) (x) = θ(x[j])θ(x[j − 1]) · · · θ(x[i + 1])θ(x[i]),
2. ((i, j), θ(x[j])),((i+1, j−1), θ(x[j−1])),. . . ,((i+1, j−1), θ(x[i+1])),((i, j), θ(x[i]))
are all inversion fragments in the i, i + 1, . . . , j − 1, j columns of Tx and have
the same center.
Note that it takes O(n2 ) time to construct Tx and the size of Tx is O(n2 ).
We also construct Ty for y.
Given a pair (((p1 , p2 ), σ1 ), ((q1 , q2 ), σ2 )) of two inversion fragments, we say
that the pair is an agreed pair if q1 = p2 + 1 or p1 + p2 = q1 + q2 . Otherwise,
we call it a disagreed pair. For two agreed pairs (((p1 , p2 ), σ1 ), ((q1 , q2 ), σ2 )) and
(((q1 , q2 ), σ2 ), ((r1 , r2 ), σ3 )), we say that two pairs are connected by ((q1 , q2 ), σ2 ).
We define an agreed sequence S to be a sequence of inversion fragments ((ai , bi ), σi )
for 1 ≤ i ≤ n, where a1 = 1, bn = n and (((ai , bi ), σi ), ((ai+1 , bi+1 ), σi+1 )) is an
agreed pair for 1 ≤ i ≤ n − 1. Note that an agreed sequence S and a set of inversions Θ has one-to-one relationship, where inversion fragments with the same
center represents the same inversion in Θ. We define an agreed sequence Sx to
be legal if there exist Θx and Θy such that Θx (x) = Θy (y) and we can generate
Sx from Θx .
The main idea of our algorithm is to keep tracking of all possible agreed pairs
for adjacent indices and check if there exist two connected pairs Px and Py for
x and y, respectively, that generate a common substring of x and y; namely,
we check if there exists a common substring σ1 σ2 σ3 and Px = (F1 , F2 ), (F2 , F3 )
for x and Py for y such that Fj yields σj . For instance, for x = AGCT and
y = CGAA, there is a common substring CT C from index 1 to 3 such that
Px = (((1, 2), C), ((1, 2), T )), (((1, 2), T ), ((3, 3)′ , C))
and
Py = (((1, 1)′ , C), ((2, 3), T )), (((2, 3), T ), ((2, 3), C))
exist.
Next, we define the following four sets for each index i:
•
•
•
•
Ah [i] = {((p, i), σ) = Tx [i][p] | 1 ≤ p ≤ i ≤ n − 1} ∪ {((i, i)′ , x[i])}.
At [i] = {((i+1, q), σ) = Tx [i+1][q +1] | i < q ≤ n}∪{((i+1, i+1)′ , x[i+1])}.
B h [i] = {((p, q), σ) = Tx [i][j] | 1 ≤ p ≤ i < q ≤ n}.
B t [i] = {((p, q), σ) = Tx [i + 1][j] | 1 ≤ p ≤ i < q ≤ n}.
Namely, Ah [i] is the set of all inversion fragments that end at i and At [i] is
the set of all inversion fragments that start from i + 1. Remark that B h [i] and
B t [i] are sets of all inversion fragments that start before i and end after i + 1.
From these four sets, we make the following observations:
Observation 2 Given a string x and its four sets Ah [i], At [i], B h [i] and B t [i],
the following statements hold:
(1) For F1 = ((p, i), σ1 ) (or ((i, i), σ1 )) in Ah [i] and F2 = ((q, r), σ2 ) ∈ B t [i],
(F1 , F2 ) is a disagreed pair. Similarly, for F1 = ((p, q), σ1 ) ∈ B h [i] and
F2 = ((i + 1, r), σ2 ) (or ((i + 1, i + 1), σ2 )) in At [i], (F1 , F2 ) is a disagreed
pair.
(2) For F1 = ((p, i), σ1 ) (or ((i, i), σ1 )) in Ah [i] and F2 = ((i + 1, q), σ2 ) (or
((i + 1, i + 1), σ2 )) in At [i], (F1 , F2 ) is an agreed pair.
(3) For F1 = ((p, q), σ1 ) ∈ B h [i] and F2 = ((r, s), σ2 ) ∈ B t [i], if p + q = r + s,
(F1 , F2 ) is an agreed pair.
Based on Observation 2, it is possible to create all agreed pairs for an index i by
comparing Ah [i] and At [i], and B h [i] and B t [i], respectively. If we can connect
pairs through all indices, then we are able to generate all agreed sequences, which
are essentially all sets of non-overlapping inversions.
We need additional tables Txc [|Σ|][|Σ|][|Σ|] and Tyc [|Σ|][|Σ|][|Σ|] to record a
sequence of three characters generated by connected pairs. For a given index i,
Txc [σ1 ][σ2 ][σ3 ] is true if there exist three inversion fragments ((ai−1 , bi−1 ), σ1 ),
((ai , bi ), σ2 )), ((ai+1 , bi+1 ), σ3 ) in a legal sequence Sx . We also define Txs [2][n + 1]
and Tys [2][n + 1] as follows. For the given index i and 1 ≤ j ≤ n + 1, Txs [1][j]
has elements (t, σ0 σ1 σ2 ) if Tx [i][j] = ((p1 , p2 ), σ1 ) is the ith element in a legal
sequence Sx , t ≤ i and ((t, p1 + p2 − t), ω) is the tth element in Sx for some ω.
Next, Txs [2][k] has elements (s, σ1 σ2 σ3 ) if Tx [i + 1][k] = ((q1 , q2 ), σ3 ) is the i + 1th
element in Sx , s ≤ i + 1 and ((s, q1 + q2 − s), τ ) is the sth element in Sx for
some τ . In other words, Txs and Tx contains information for the ith agreed pair
in legal Sx .
We design an algorithm that computes Txs and Tys for each index and checks
whether or not Txs and Tys are empty for the last index. If Tx [1][j] yields σ, we
set Txs [1][j] = {(1, AAσ)} as initial data. We also set Tys similarly. Now we run
the following steps from index 1 to n − 1 for x and y. We only illustrate the case
for x. (The case for y is similar.)
Step 1. We check all inversion fragments in ith column of Tx . For Tx [i][j] =
((j, i), σ3 ) (or ((i, i)′ , σ3 )), if (j, σ1 σ2 σ3 ) ∈ Txs [1][j], then we add ((j, i), σ2 σ3 ) (or
((i, i)′ , σ2 σ3 )) to a set Ah . Namely, Ah contains every inversion fragment that
can be the ith element in a legal sequence and ends at i.
Step 2. We check all inversion fragments in i + 1th column of Tx . For Tx [i +
1][i + 1] = ((i + 1, i + 1),′ σ), we add ((i + 1, i + 1)′ , σ) to a set At . Moreover, for
each Tx [i + 1][j] = ((i + 1, j − 1), σ), we add ((i + 1, j − 1), σ) to a set At . In other
words, At contains every inversion fragment that can be the i + 1th element in
a legal sequence and starts from i + 1.
Lemma 1. Each cell in Txs and Tys has O(n) elements.
Proof. Elements in Txs and Tys are of the form (p, σ1 σ2 σ3 ). The number of the
combination for the second part is |Σ|3 , which is constant. For the first part,
1 ≤ p ≤ i for index i since p is the start index of the last inversion in an inversion
sequence. Therefore, the number of possible elements for each cell is O(n). ⊓
⊔
Lemma 2. We can build Ah and At in O(n2 ) using O(n) space.
Proof. For Step 1, we need to check i + 1 inversion fragments, and for each
inversion fragment we need to check O(n) elements by Lemma 1. Therefore, we
need O(n2 ) for Step 1 and Ah has at most i + 1 inversion fragments. For Step 2,
At has n − i inversion fragments. Therefore, the total process takes O(n2 ) time
and requires O(n) space to store all inversion fragments.
⊓
⊔
Once we have two set of inversion fragments representing Ah [i] and At [i], we
can calculate all agreed pairs generated from Ah [i] and At [i].
Step 3. Based on Observation 2(2), for every ((p1 , i), σ1 σ2 ) (or ((i, i)′ , σ1 σ2 )) in
Ah and ((i + 1, p2 ), σ3 ) (or ((i + 1, i + 1)′ , σ3 )) in At , we set Txc [σ1 ][σ2 ][σ3 ] = true.
We also add (i + 1, σ1 σ2 σ3 ) to Txs [2][p2 ].
Lemma 3. Given Ah and At , we can update Txc and Txs in O(n2 ) time by Step 3.
Proof. Since |Ah | ≤ i + 1 and |At | = n − i, we repeat Step 3 at most (i + 1)(n − i)
times. Thus, we can update Txc and Txs in O(n2 ) time.
⊓
⊔
We have calculated all agreed pairs generated from Ah [i] and At [i] in Step 3,
and the other case of generating agreed pairs for the index i is to use the inversion
fragments with same center from B h [i] and B t [i]. Note that inversions with same
center represent the same inversion from Observation 1.
Step 4. Based on Observation 2(3), for ((p1 , p2 ), σ2 ) = Tx [i][j] and ((q1 q2 ), σ3 ) =
Tx [i+1][k], where p1 +p2 = q1 +q2 and i ̸= j and i+1 ̸= k, if (t, σ0 σ1 σ2 ) ∈ Txs [1][j]
and t ≤ p1 , then we set Txc [σ1 ][σ2 ][σ3 ] = true and add (t, σ1 σ2 σ3 ) to Txs [2][k].
Lemma 4. We can update Txc and Txs in O(n2 ) time by Step 4.
Proof. As illustrated in Fig. ??, we can construct Tx such that all inversion
fragments at each column are sorted by the center. Therefore, we can check two
inversion fragments with same center in O(n). Since for each inversion fragment
we need to check O(n) elements in Txs from Lemma 1, the whole process takes
O(n2 ).
⊓
⊔
Step 5. For all three letter strings σ1 σ2 σ3 over Σ,
{
true if Txc [σ1 ][σ2 ][σ3 ] and Tyc [σ1 ][σ2 ][σ3 ] are both true,
c
Tx [σ1 ][σ2 ][σ3 ] =
false otherwise.
Once we recompute Txc , then, for each (p, σ1 σ2 σ3 ) in Txs , we remove (p, σ1 σ2 σ3 )
from Txs if Txc [σ1 ][σ2 ][σ3 ] is false. The process ensures that Txs and Tys produce
the same sequence of characters by connected pairs.
Lemma 5. It takes O(n2 ) time to generate Txs and Tys such that elements in
both array generate the same set of sequences of characters.
Proof. After modifying Txc , we need to remove elements in Txs according to Txc .
Since the size of Txs is O(n2 ) and the size of Txc is constant, the process takes
O(n2 ) time.
⊓
⊔
We are now ready to present the whole procedure of our algorithm. (Algorithm 1 is a pseudo description of the proposed algorithm.)
Algorithm 1:
1
2
3
4
5
6
7
8
9
10
11
12
Input: String x, y
Output: Boolean (whether or not there exists Θx , Θy s.t. Θx (x) = Θy (y).)
/* time complexity: O(n3 ), space complexity: O(n2 )
make Tx and Ty .
initialize Txs and Tys .
for i ← 1 to n − 1 do
clear and initialize arrays used in previous iteration.
store information for agreed sequences to Txs , Txc , Tys , Tyc .
Txc , Tyc ← Txc ∧ Tyc
modify Txs and Tys according to Txc and Tyc .
copy the second column of Txs and Tys to the first column.
*/
if the second column of Txs and Tys are not empty then
return true
else
return false
Once we finish calculating Txs and Tys using Steps 1, 2, 3, 4 and 5 from index
1 to n − 1, we check whether or not the last columns in Txs and Tys are empty. If
they are not empty, then there are agreed sequences for x and y that generate
the same string, which means that x and y can generate the same string by
applying non-overlapping inversions. If they are empty, then there is no agreed
sequences for x and y that generate the same string. The proposed algorithm
runs in O(n3 ) time using O(n2 ) space.
Theorem 3. We can solve the the the alignment with non-overlapping inversion
problem in Definition 1 in O(n3 ) time using O(n2 ) space, where n is the size of
input strings.
4
Conclusions
The inversion is an important operation for bio sequences such as DNA or RNA
and is closely related to mutations. We have, in particular, considered nonoverlapping inversions on both bio sequences, which is important to find the
original common sequence from two mutated sequences. We have proposed a
new problem, alignment with non-overlapping inversions on two strings, and
presented a polynomial algorithm for the problem. Given two strings x and y,
based on the inversion properties, our algorithm decides whether or not there
exist two sets Θx and Θy of inversions for x and y such that Θx (x) = Θy (y)
in O(n3 ) time using O(n2 ) space, where n = |x| = |y|. The proposed problem
is about the sequence alignment and can be extended to approximate pattern
matching or edit distance problem.
References
1. D. Cantone, S. Cristofaro, and S. Faro. Efficient string-matching allowing for nonoverlapping inversions. Theoretical Computer Science, 483:85–95, 2013.
2. D. Cantone, S. Faro, and E. Giaquinta. Approximate string matching allowing for
inversions and translocations. In Proceedings of the Prague Stringology Conference,
pages 37–51, 2010.
3. Z.-Z. Chen, Y. Gao, G. Lin, R. Niewiadomski, Y. Wang, and J. Wu. A spaceefficient algorithm for sequence alignment with inversions and reversals. Theoretical
Computer Science, 325(3):361–372, 2004.
4. S. Grabowski, S. Faro, and E. Giaquinta. String matching with inversions and
translocations in linear average time (most of the time). Information Processing
Letters, 111(11):516–520, 2011.
5. Z. Ignatova, K. Zimmermann, and I. Martinez-Perez. DNA Computing Models.
Advances in Information Security, v. 42. 2008.
6. J. D. Kececioglu and D. Sankoff. Exact and approximation algorithms for the
inversion distance between two chromosomes. CPM ’93, pages 87–105, 1993.
7. S. C. Li and Y. K. Ng. On protein structure alignment under distance constraint.
ISAAC ’09, pages 65–76, 2009.
8. O. Lipsky, B. Porat, E. Porat, B. R. Shalom, and A. Tzur. Approximate string
matching with swap and mismatch. ISAAC’07, pages 869–880. 2007.
9. C. M. Ogilvie and P. N. Scriven. Meiotic outcomes in reciprocal translocation carriers ascertained in 3-day human embryos. European Journal of Human Genetics,
10(12):801–806, 2009.
10. M. Oliver-Bonet, J. Navarro, M. Carrera, J. Egozcue, and J. Benet. Aneuploid
and unbalanced sperm in two translocation carriers: evaluation of the genetic risk.
Molecular Human Reproduction, 8(10):958–963, 2002.
11. T. S. Painter. A New Method for the Study of Chromosome Rearrangements and
the Plotting of Chromosome Maps. Science, 78:585–586, Dec. 1933.
12. Y. Sakai. A new algorithm for the characteristic string problem under loose similarity criteria. ISAAC’11, pages 663–672, 2011.
13. M. Schöniger and M. S. Waterman. A local algorithm for DNA sequence alignment
with inversions. Bulletin of Mathematical Biology, 54(4):521–536, 1992.
14. A. F. Vellozo, C. E. R. Alves, and A. P. do Lago. Alignment with non-overlapping
inversions in O(n3 )-time. WABI’06, pages 186–196, 2006.