MSc in High Performance Computing MSc in High
advertisement

MSc in High Performance Computing
MSc in High Performance Computing with Data Science
An introduction to integrated development environments
and Eclipse
Mike Jackson
January 8, 2016
1
Introduction
An integrated development environment, or IDE, provides a comprehensive tool suite for software development. An IDE typically includes a source code editor, compiler or interpreter, a debugger and a file
browser. Many support additional features such as build tools, support for running and reporting on tests,
integrated API documentation browsing, revision control support etc.
There are many different integrated development environments available. Three popular, and free, IDEs
are:
• Eclipse. Primarily for Java with plugins available for C/C++, FORTRAN, Python, PHP, Perl,
JavaScript, Python, Ruby and R.
• NetBeans for C/C++, HTML, PHP and Java.
• Microsoft Visual Studio Express. Primarily for C/C++, C# and VB.NET with plugins available for
JavaScript and Ruby.
Though these differ in the specific features they support, even within different versions of the same
product, the basic principles are the same.
In this walkthrough, we’ll be using Eclipse 4.2 Juno, installed on the Physics Computational Lab machines. Though some Java knowledge may help, the primary aim of the walkthrough is to demonstrate
the features of an IDE.
2
DNA sequences
We will be developing code and tests that implement operations on DNA sequences. This example is
inspired originally by the Python DNA function of Gordon Webster, the Digital Biologist1 ).
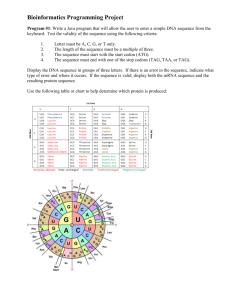
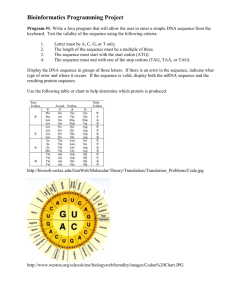
A DNA sequence is a chain of one or more organic molecules called nucleotides. Each nucelotide adenine, cytosine, guanine, thymine - is represented by a letter - A, C, G and T. A DNA sequence is a
chain of these e.g. AAAGTCTGAC.
Each of these nucleotides has a molecular weight:
1 http://www.digitalbiologist.com/2011/04/code-tutorial-getting-started-with-python.html
1
• Adenine (A): 131.2
• Cytosine (C): 289.2
• Guanine (G): 329.2
• Thymine (T): 304.2
The molecular weight of a DNA sequence is the sum of the molecular weights of each of its nucleotides.
For example, the molcular weight of GATTACCA is:
• A: 131.2 * 3 = 393.6
• C: 289.2 * 2 = 578.4
• G: 329.2 * 1 = 329.2
• T: 304.2 * 2 = 608.4
• Molecular weight: 1909.6
3
Create a new Eclipse project
Open a shell window and type:
$ eclipse-4.2
The Workspace Launcher will appear. Check that you are happy with the proposed path to a new
workspace directory in your home directory, then click OK.
The Eclipse interface will appear.
Click the X on the Welcome area, or view, to free up some screen space.
We will now create a new project for our code.
• Select File => New => Java Project
• Enter Project name: DNA
• Click Finish
Our project will be created and the Java perspective will appear. A perspective in Eclipse provides a
related sets of areas, or views and editors.
On the left is the Package Explorer, which allows us to browse our source code. In the centre is where
our source code editor will appear. Below this are various status and information views.
4
Create a Java package
We will create a new Java package. Java packages provide us with a way to collect related classes
together.
• In the Package Explorer, click the + beside DNA
• Right-click on src and select New => Package
• In the New Java Package dialog, enter Name: dna
• Click Finish
In the Package Explorer, the dna package will appear.
2
5
Create a class
We will create a new class, Sequence, to represent our DNA sequences.
• In the Package Explorer, right-click on the dna package and select New => Class
• In the New Java Class dialog, enter Name: Sequence
• Click Finish
In the Package Explorer, the Sequence.java source code will appear under the dna package, and a
new editor will appear with some “boiler-plate” code - a template for our class.
Now enter the following code.
package dna;
import java.util.HashMap;
public class Sequence {
/** Map from nucleotides to molecular weights. */
public static final HashMap<Character, Double> WEIGHTS;
/** Nucleotides in the sequence. */
private String nucleotides;
/** Initialise the WEIGHTS map. */
static {
WEIGHTS = new HashMap<Character, Double>();
WEIGHTS.put(’A’, 131.2);
WEIGHTS.put(’C’, 289.2);
WEIGHTS.put(’G’, 329.2);
WEIGHTS.put(’T’, 304.2);
}
public Sequence(String nucleotides) {
this.nucleotides = nucleotides;
}
public String getNucleotides() {
return this.nucleotides;
}
public double getWeight() {
return calculateWeight(this);
}
}
Look at the Outline view on the right. This lists the package and class structure e.g. constants, attributes
and methods.
Any errors are highlighted with an X and a red line. If you move the mouse over these, you’ll be told what
the problem is. Here the problem is that ’The method calculateWeight(Sequence) is undefined
for the type Sequence’.
Eclipse can help solve the error, by doing a “quick-fix” for you:
• Click on the X to the left of calculateWeight
• Double-click ’Create method calculateWeight(Sequence)’
A method skeleton will be created:
private double calculateWeight(Sequence sequence) {
// TODO Auto-generated method stub
return 0
}
3
Let’s change it to be a public static method:
public static double calculateWeight(Sequence sequence) {
Now, provide the method body. Enter the following and stop after the “.”
char[] chars = sequence.
If you type an object reference e.g. sequence. when you pause at the “.” a list of methods supported by
the object appears.
Double-click on getNucleotides and it will be inserted. This is auto-completion.
Now enter a . after this and again a list will be shown. This time, though, you can also see the inputs and
outputs of each method and a description of its behaviour. This is because the Java API documentation
is available. We’ll see how we can do this for our own code shortly.
Double-click on toCharArray() to insert it.
Add a final ; to give:
char[] chars = sequence.getNucleotides().toCharArray();
Now add in:
double weight = 0;
Any problems which aren’t errors but which may be of concern e.g. local variables that are never read,
are highlighted with a warning - a yellow triangle and an exclamation mark. If you move the mouse over
these and click them, you’ll be told what the problem is.
Now finish the method by adding:
for (char c: chars) {
weight += WEIGHTS.get(c);
}
return weight;
Delete the line:
return 0;
And delete the comment:
// TODO Auto-generated method stub
as we have now implemented our method.
Eclipse will regularly try and build our project. You can see this if you go, in a shell window, to your
Eclipse workspace directory, where you should see a bin/ directory with the classes:
$ ls -R workspace/DNA/bin
workspace/DNA/bin/:
dna
workspace/DNA/bin/dna:
Sequence.class
Click on the Project menu and you’ll see that Build Automatically is enabled. For large projects with lots
of code you might want to turn this off and only build it when you’re ready.
4
6
Write JavaDoc
JavaDoc is a way of writing comments for Java which are machine-readable. It’s a markup notation
for commenting code which can be parsed and from which documentation can be automatically generated. From JavaDoc we can create a set of HTML pages, all cross-referenced and hyper-linked, which
can be a useful resource for other developers. For example see the Apache Axis2/Java JavaDoc at
http://axis.apache.org/axis2/java/core/ - click the Online Java Docs link on the left (the direct link is
http://axis.apache.org/axis2/java/core/api/index.html).
In Eclipse, JavaDoc is parsed to offer information when doing auto-completion.
Move the cursor to the line before:
public static double calculateWeight(Sequence sequence) {
and type:
/**
which is the markup to open up a comment block. Press return. Eclipse parses the method and creates a
template for the comments for us. Note the @param and @return JavaDoc tags.
/**
*
* @param sequence
* @return
*/
We can now fill this in. For example:
/**
* Calculate the molecular weight of a DNA sequence.
* @param sequence DNA sequence.
* @return molecular weight formed by summing the weight of each
* nucleotide in turn.
*/
Now add JavaDoc for getWeight e.g.
/**
* Get the molecular weight of this DNA sequence.
* @return molecular weight formed by summing the weight of each
* nucleotide in turn.
*/
Now start to create a main method, the first method that will be called when we run our program:
public static void main(String[] arguments) {
Sequence sequence = new Sequence(arguments[0]);
Now enter the following and stop at the “.”
System.out.println(sequence.
and scroll down and click getWeight. You’ll now see your JavaDoc.
Double-click on getWeight and add );
System.out.println(sequence.getWeight());
so that the main method is:
5
public static void main(String[] arguments) {
Sequence sequence = new Sequence(arguments[0]);
System.out.println(sequence.getWeight());
}
7
Run the code
Our class has a main method which is Java’s entry point for running a program. We can run our code
by clicking on the run icon - it is a green circle with a white triangle. Eclipse now acts as if we’d run our
code from the command-line. A Console view appears in which we can see we have a problem. This is
because our program expects us to provide a DNA sequence as an argument So:
• Click on the “V” in the run icon - it is a green circle with a white triangle
• Select Run Configurations...
• Click Sequence under Java Application
• Click on the Arguments tab
• In Program arguments enter: GATTACCA
• Click Run
In the Console we should now see:
1909.6000000000001
Note the rounding error - computers don’t do floating point arithmetic too well! We’ll return to this
shortly.
Click on the run icon again and it’ll run the same configuration as last time i.e. using our command-line
argument.
8
Debug our program
When we ran our program we used the run icon. Next to this is the debug icon, signified by a green bug.
Click this icon and the Console should show the same value as when we clicked on the run icon:
1909.6000000000001
To the left of the line
public static double calculateWeight(Sequence sequence) {
right-click and select Toggle Breakpoint. A little blue blob will appear.
Now click the Debug icon again.
A Confirm Perspective Switch dialog will appear as Eclipse asks to switch from the Java perspective
(remember that a perspective is a collection of views and editors) to the Debug perspective. Click Yes
and Eclipse will change perspective, putting up views associated with debugging.
In the Debug view you should see that execution has paused at Sequence.calculateWeight,
where we set the breakpoint.
The Sequence.java editor, which may have changed location on screen, will have this line highlighted (you may have to scroll the editor window to see it).
6
A Variables view shows the name of currently declared variables in this case sequence. If the variable
is an object, as this one is, you can double-click on it and see its attributes.
Double-click sequence and see the value of nucleotides. It should be GATTACCA.
There will be a number of additional available, represented as arrow-related icons in the toolbar, to control
what happens next including:
• Resume - carry on execution until the next breakpoint is reached.
• Terminate - stop everything!
• Step into - this allows you to dive into the execution of a specific line e.g. into the commands
within any method called within the current line.
• Step over - this executes the current line and moves onto the next line in the current control flow.
• Step return - allow the current method to execute to completion then stop at the line which called
it.
Hover the mouse over each icon to find out which is which.
Click Step over repeatedly (or press F6) and watch how the current line repeatedly enters the loop and,
in the Variables view, how the value of the weight variable increases as each nucleotide’s molecular
weight is added to weight in turn.
If you’re tired of this you can see what happens if you click Step return (or press F7).
If you want to continue execution to completion, click Resume, the green right-pointing arrow (or press
F8).
To get back to the Java perspective select Window => Open Perspective => Java.
9
Export the compiled code to a JAR file
We can export our code as one (or more) JAR file(s) (Java ARchive files), if we want to distribute it. JAR
files are libraries of compiled Java classes. They are similar to libraries in C or Fortran, though are more
portable, in that they can run on any machine that has a Java Runtime Environment - a JAR file created
under Linux can be used under Windows, for example. To export our code as a JAR file:
• Right-click on src under DNA in the Package Explorer.
• Select Export...
• Double-click Java
• Select JAR file
• Click Next
• You can choose what parts of your project to bundle into the JAR file. We’ll use the defaults for
now.
• Click Browse... and enter Name: dna.jar and select the directory where you want your JAR file to
go.
• Click OK
• Click Finish
In a shell window, look for your JAR.
Run the Java program. To do this we need to specify our CLASSPATH - in this case dna.jar - the
class with our main method - dna.Sequence - and any required command-line arguments - our DNA
sequence e.g.
7
$ java -classpath dna.jar dna.Sequence GATTACCA
1909.6000000000001
The -classpath argument specifies a list of paths to JAR files in the file system where all the classes
we need can be found. Our dna.Sequence class is in dna.jar.
Java will look for the class dna.Sequence in dna.jar and, if it finds it, will look for a main method
in that class. This serves as the entry point for our program and Java will then execute our program
starting with our main method.
10
Write and run unit tests
We’ll now write some tests for our class. Here, we focus on how Eclipse helps us write and run tests.
First we need to include JUnit (http://junit.org), the Java unit text framework, in our available libraries:
• Right-click on DNA in the Package Explorer and select Properties.
• Double-click Java Build Path
• Click Libraries tab
• Click Add Library...
• Select JUnit
• Click Next
• Select JUnit library version: JUnit 4
• Click Finish
• Click OK
It is useful to keep tests separate from code as when building code for release we don’t want one bloated
binary file with all the code and tests but rather to keep these separate - users can get the code they need,
and developers can get the tests:
• Right-click on DNA in the Package Explorer and select New => Source Folder.
• Enter Folder name: test
• Click Finish
Create a dna package within test, using the same approach to create a package that you used earlier.
Now:
• Right-click on test in the Package Explorer and select New => JUnit Test Case
• Enter Package: dna
• Enter Name: SequenceTest
• Click Browse... next to Class under test:
• Enter Sequence and under Matching items select Sequence - dna - DNA/src
• Click OK
• Click Next
• Select for what methods we want test methods created. Check:
– getNucleotides()
8
– getWeight()
– calculateWeight(Sequence)
• Click Finish
A new editor will appear with our test class, SequenceTest.java. Note how there are test methods
already provided (denoted by the @Test annotation) and that they are by default set to fail, as a reminder
that we need to implement them!
Let’s run our tests. We can run our test class by clicking on the run icon - it is a green circle with a white
triangle.
A JUnit view appears giving us a report on which tests succeeded and which failed. As ours are all set to
fail they do indeed fail!
If you click on a method in the JUnit view the cause of failure is shown in the Failure Trace part of the
view.
Here we ran the tests in a single class. We can run every test in the dna package as follows:
• Select test in the Package Explorer
• Click on the “V” in the run icon
• Select Run As => JUnit Test
This will run the tests in all classes, though, as we have only one test class it looks the same as before!
Alternatively we can run all the tests in a class, or even a specific test in a class via the Outline view:
• Right-click on testGetWeight in Outline view, Package explorer or in the source code editor
• Select Run As => JUnit Test
Let’s implement three tests. First we need to import our Sequence class so add:
import dna.Sequence;
It will be hidden to save screen space, so click the “+” next to the line
import static org.junit.Assert.*;
to see it again.
Let’s write a test to check that if we create a Sequence with a given string of nucelotides, that getNucleotides
returns us that string:
@Test
public void testGetNucleotides() {
String sequenceStr = "GATTACCA";
Sequence sequence = new Sequence(sequenceStr);
assertEquals("Nucleotides returned were not those given",
sequenceStr, sequence.getNucleotides());
}
Now, if we run our test class by clicking on the run icon, we should get one success and two failures.
Add in the following tests for getWeight and calculateWeight:
@Test
public void testGetWeight() {
Sequence sequence = new Sequence("G");
assertEquals("Weight returned was unexpected",
9
Sequence.WEIGHTS.get(’G’).doubleValue(), sequence.getWeight());
}
@Test
public void testCalculateWeight() {
Sequence sequence = new Sequence("G");
assertEquals("Weight returned was unexpected",
Sequence.WEIGHTS.get(’G’).doubleValue(),
Sequence.calculateWeight(sequence));
}
We validate the weight of G against that stored in WEIGHTS rather than hard-coding the value 289.2 in
our test class. This means that if we need to change our value of G we only need to do so in one place.
10.1
Getting round floating-point problems
Note that assertEquals has been struck-through. This is a warning that we’re using a deprecated
method which may be removed from a later version of JUnit.
assertEquals is polymorphic and comes in many versions. The version we use in testGetNucleotides
compares two doubles for equality. It has three arguments:
1. Informative message for if the test fails.
2. Expected double, our expected value.
3. Actual double, our actual value, the value from our method that we are testing for correctness.
The version we should use in testGetWeight, testCalculateWeight takes four arguments:
1. Informative message for if the test fails.
2. Expected double, our expected value.
3. Actual double, our actual value, the value from our method that we are testing for correctness.
4. A delta.
The delta allows us to get round the problem of floating point numbers we saw earlier. Instead of comparing the numbers for exact equality, it compares them to equality within the threshold determined by
this delta i.e. it asserts true if
Math.abs(expected - actual) < delta
So let’s add 0.01 as our delta in each of these calls to assertEquals and rerun the tests. The choice
of an appropriate delta depends very much on the nature of the application being tested.
10.2
Export the compiled tests to a JAR file
Let’s export the tests into a new JAR file. Start by right-clicking on test in the Package Explorer. Call
your JAR file dna-test.jar.
In a shell window, download a JUnit library from http://junit.org.2
In a shell window, look for your dna-test.jar JAR then run the following. Do not type the \
character (this is a character commonly used to indicate that the following line is to be treated as part of
the current line) - type the whole command on a single line:
2A
direct link is http://search.maven.org/remotecontent?filepath=junit/junit/4.10/junit-4.10.jar
10
$ java -classpath dna.jar:dna-test.jar:junit-4.10.jar \
org.junit.runner.JUnitCore dna.SequenceTest
You should see:
JUnit version 4.10
....
Time: 0.007
OK (3 tests)
The -classpath argument specifies a list of paths to JAR files in the file system where all the classes
we need can be found. These are separated by :. Our dna.Sequence class is in dna.jar, our test
class is in dna-test.jar, and JUnit’s classes are in junit-4.10.jar.
The Java program we are running is org.junit.runner.JUnitCore, the JUnit test runner. The
argument to this program is our test class, dna.SequenceTest. The JUnit test runner will look for
test methods in our test class and will run any it finds.
11
Importing code into Eclipse projects
You may have code that you want to pull into an Eclipse project. You can do this as follows:
• Double-click project name, in Package Explorer
• Right-click src
• Select Import...
• Double-click General
• Double-click File system
• Next to From Directory,. Click Browse...
• Browse to the directory containing the Java files you want to import
• Click OK
• Select the Java files
• Click Finish
This will copy the Java files into your project workspace and you will be able to edit, compile, and run
them as above.
12
Using Eclipse with external projects
A common question is “how does one use Eclipse with code that has been developed outside Eclipse?”
This can occur if, for example, you are in a team developing software that can be built and tested automatically (so, for example, provides ANT scripts to build and test the software) but also gives the team
members the flexibility to choose their own IDE whether that be Eclipse or even a text editor and the
command-line Java Development Kit tools. Assuming your source code is held in version control, you
don’t want to import your code into the workspace (as described in the previous section) as this copies the
imported code. Rather, you want to leave the code where it is (for example in a clone of a Git repository,
or a checkout of a Subversion repository) in your file system.
To see how this works, in a shell window, run:
11
$ git clone https://github.com/softwaresaved/build_and_test_examples
This clones a Git repository with code samples and build scripts for a number of examples. Now change
into the java directory:
$ cd build_and_test_examples/java
If you explore this directory you will see it has src/ and test/ directories, each with a math package,
classes and, in test/, JUnit test classes. Now, if we wanted to develop this code and run the tests via
Eclipse we can do this as follows.
Create a new Java Project:
• Select File => New => Java Project
• Enter Project name: MathProject
• Click Finish
Delete the default src/ directory Eclipse creates:
• Under MathProject, in Package Explorer, right-click src
• Select Delete
Link to your Java soource code folder, e.g. build_and_test_examples/java/src:
• Right-click MathProject in Package Explorer
• Select Build Path => Link Source...
• Click Browse...
• Browse to your source code folder e.g. build_and_test_examples/java/src
• Click OK
• Click Finish
Link to your directory’s Java test code folder e.g. build_and_test_examples/java/test:
• Right-click MathProject in Package Explorer
• Select Build Path => Link Source...
• Click Browse...
• Browse to your test folder e.g. build_and_test_examples/java/test
• Click OK
• Click Finish
Add the JUnit library to the project:
• Right-click MathProject in Package Explorer
• Select Properties
• Double-click Java Build Path
• Click Libraries tab
• Click Add Library...
• Select JUnit
• Click Next
12
• Select JUnit library version: JUnit 4
• Click Finish
• Click OK
Linked directories are held as absolute paths in the Eclipse configuration. If you move the directory
containing your src/ and test/ directories (e.g. you check it out of a repository onto another machine) then
you need to update the paths. You can change these as follows:
• Right-click project-name
• Select Build Path => Configure Build Path
• Click Source tab
You can then edit the paths to point to the new locations.
Or, you can edit your Eclipse configuration manually:
• Exit Eclipse
• Open workspace/PROJECTNAME/.project in an editor
• Edit the paths. These are in a linkedResources section e.g.
<linkedResources>
<link>
<name>src</name>
<type>2</type>
<location>/home/user/build_and_test/java/src</location>
</link>
<link>
<name>test</name>
<type>2</type>
<location>/home/user/build_and_test/java/test</location>
</link>
</linkedResources>
• Restart Eclipse.
13
Conclusion
That concludes the walkthrough of Eclipse. The key points to take away are that Eclipse and many other
IDEs provide a lot of tools to take away some of the grunt-work in writing code, writing comments,
writing tests, running tests and building binaries, allowing a developer to spend more of their effort on
the intellectually challenging aspects of writing code!
13