Family of Bears Phylogenetic Tree
advertisement

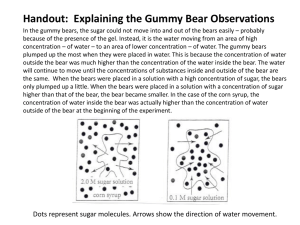
NAME: ____________________________________ DATE: ______________ BLOCK: ____ Are polar bears closely related to American brown bears? Do you think the giant panda is more closely related to the American brown bear or the polar bear? Is the giant panda a bear at all? What about the red panda – is it a giant panda relative? What does the evolutionary history of bears look like? In this activity you will use real gene sequences to figure out which animals are more closely related. You will create a family tree for bears and help to solve the mystery of bear history! TABLE 1: Accession numbers and scientific names different species 1. Organism Accession Number American black bear Y08520 American brown bear L21889 Spectacled bear L21883 Asiatic bear L21890 Polar bear L22164 Giant panda Y08521 Red Panda S80939 Northern raccoon U78345 Leopard seal AY377288 Scientific Name Go to NCBI (National Center for Biotechnology Information) www.ncbi.nlm.nih.gov. To the left of the Search button are (1) a list Drop-down List box and (2) an Edit box. In the drop-down List box, select Nucleotide. In the center Edit box, paste the Accession Number (from Table 1 above) for the American Black bear. Click the Search button. 2. When the entry pops up, you’ll see the following: U. americanus mitochondrial 12S rRNA gene. U. americanus is the scientific name for the American black bear. The search found a partial gene sequence for this species’ mitochondrial DNA that makes up part of each ribosome. Enter the scientific name in the 3rd column of Table 1. 1 3. Click the FASTA link near the top of the page to view the DNA sequence. Highlight and copy the entire sequence including the title (everything from the > onward). >gi|1871572|emb|Y08520.1| U.americanus mitochondrial 12S rRNA gene CAAAGGTTTGGTCCTGGCCTTCCTATTAGCTGCTAACAAGATTACACATGTAAGTCTCCGCGCCCCAGTG AAAATGCCCTTTGGATCTTAAAGCGGTTCGAAGGAGCGGGCATCAAGCACACCTCTCCCCGGTAGCTTAT AACGCCTTGCTTAGCCACACCCCCACGGGATACAGCAGTGATAAAAATTAAGCTATGAACGAAAGTTTGA CTAAGCTATGTTGATCAAAGGGTTGGTTAATTTCGTGCCAGCCACCGCGGTTATACGATTGACCCAAGTT AATAGGCCCACGGCGTAAAGCGTGTGAAAGAAAAATTTTTTCCCCACTAAAGTTAAAGTTTAACCAAGCT GTAAAAAGCTATCGGTAACACTAAAATAAACTACGAAAGTGACTTTAATACTCTCAACCACACGACAGCT AAGATCCAAACTGGGATTAGATACCCCACTATGCTTAGCCTTAAACATAAGTAATTTATTAAACAAAATT ATTCGCCGGAGAACTACTAGCAACAGCTTAAAACTCAAAGGACTTGGCGGTGCTTTAAACCCCCCTAGAG GAGCCTGTTCTATAATCGATAAACCCCGATAGACCTCACCACCTCTTGCTAATCCAGTCTATATACCGCC ATCTTCAGCTAACCCTTAAAAGGAGTAAAAGTAAGCACAATCATCCCACATAAAAAAGTTAGGTCAAGGT GTAACCCATGGGGTGGGAAGAAATGGGCTACATTTTCTATTCAAGAACAACCTACGAAAGTTTTTATGAA ACTAAAAACTAAAGGTGGATTTAGCAGTAAACCAAGAATAGAGAGCTTGGTTGAATAGGGCAATGGAGCA TGCACACACCGCCCGTCACCCTCCTCAAGTGGCACAAGTCAAATATAACCTATTGAAATTAAATAAAACG CAAGAGGAGACAAGTCGTAACAAGGTAAGCATACTGGAAAGTGTGCTTGGATAAAC 4. Open an MS-Word document and call it Fasta.docx. Paste the sequence into the document. Change the Title line (the first line, it begins with >) to: >American-black-bear-Y08520. There should be NO SPACES anywhere on the line. 5. Collect the DNA sequences for all 9 species, and change the title lines in a similar fashion. 6. In a 2nd browser tab, go to the Next Generation Biology Workbench page (http://www.ngbw.org/) User: ______________________ 7. Click Use the Workbench now. PW: _______________________ Click on Create an account. Fill out the 7 required fields, and click Register. Enter your password and user name in the box above so that you do not forget it. 8. Log in to your account. Click Create New Folder, call it Bears, and click on Save. Two subfolders are automatically created: Data and Tasks. Click on Data. 2 9. Click on Upload Files. You will not be Uploading files – rather you'll be pasting your data into the browser, as follows: In the Label field, paste the Title (without the >), e.g.: American-black-bear-Y08520 In the box where it says: Enter your data, paste in the entire record, including the title line, e.g.: >American-black-bear-Y08520 CAAAGGTTTGGTCCTGGCCTTCCTATTAGCTGCTAACAAGATTACACATGTAAGTCTCCGCGCCCCAGTGAAAATGCCC TTTGGATCTTAAAGCGGTTCGAAGGAGCGGGCATCAAGCACACCTCTCCCCGGTAGCTTATAACGCCTTGCTTAGCCAC ACCCCCACGGGATACAGCAGTGATAAAAATTAAGCTATGAACGAAAGTTTGACTAAGCTATGTTGATCAAAGGGTTGGT TAATTTCGTGCCAGCCACCGCGGTTATACGATTGACCCAAGTTAATAGGCCCACGGCGTAAAGCGTGTGAAAGAAAAAT TTTTTCCCCACTAAAGTTAAAGTTTAACCAAGCTGTAAAAAGCTATCGGTAACACTAAAATAAACTACGAAAGTGACTT TAATACTCTCAACCACACGACAGCTAAGATCCAAACTGGGATTAGATACCCCACTATGCTTAGCCTTAAACATAAGTAA TTTATTAAACAAAATTATTCGCCGGAGAACTACTAGCAACAGCTTAAAACTCAAAGGACTTGGCGGTGCTTTAAACCCC CCTAGAGGAGCCTGTTCTATAATCGATAAACCCCGATAGACCTCACCACCTCTTGCTAATCCAGTCTATATACCGCCAT CTTCAGCTAACCCTTAAAAGGAGTAAAAGTAAGCACAATCATCCCACATAAAAAAGTTAGGTCAAGGTGTAACCCATGG GGTGGGAAGAAATGGGCTACATTTTCTATTCAAGAACAACCTACGAAAGTTTTTATGAAACTAAAAACTAAAGGTGGAT TTAGCAGTAAACCAAGAATAGAGAGCTTGGTTGAATAGGGCAATGGAGCATGCACACACCGCCCGTCACCCTCCTCAAG TGGCACAAGTCAAATATAACCTATTGAAATTAAATAAAACGCAAGAGGAGACAAGTCGTAACAAGGTAAGCATACTGGA AAGTGTGCTTGGATAAAC Configure the Data Type fields as follows: Entity Type: Nucleic Acid (RNA or DNA) Data Type: Sequence Data Format: FASTA (.fsa, .fasta, .fst) Click Save. Click Return to Data List. Enter all 9 DNA sequences in a similar way. 10. Click on the Tasks subfolder. Click Create New Task. Name the task: Bears-Tree. Click Set Description. 11. Click Select Input Data. Click Select all to check all 9 DNA sequences. Click Add Selected to Task. 12. Click Select Tool. Select / click ClustalW-N. 13. Click Save and Run Task. 14. Wait until View Status turns into View Output. Click View Output 3 15. To see how the sequences aligned (lined up), click View File for either outfile.aln entry. . 16. To see the Phylogenetic Tree, click View File for either newtree.dnd entry. Click Save to Current Folder. Select the Type of Data parameters as follows: Entity Type: Taxon Data Type: Phylogenetic Tree Data Format: Newick (Phylip) Tree (.dnd) Click Save. Click on the Data folder. Click on the tab Phylogenetic Tree. Click on the Label: newtree.dnd. Click Draw Tree. 4