5 Introduction to DNA sequence analysis
advertisement

MAS3326/8326 Discrete Stochastic Modelling
5
Introduction to DNA sequence analysis
5.1
Background and motivation
This part of the course is all about using probability theory to construct models for DNA
sequences, and then using statistical techniques to infer the parameters of these models.
First, let’s take a look at some background information about DNA sequences.
• A DNA sequence is a string of nucleic acids attached to a sugar-phosphate backbone.
• There are four types of nucleic acid or base, namely adenine (A), cytosine (C),
guanine (G) and thymine (T).
• Sequences possess an orientation due to the way they are attached to the chemical
backbone.
• Because of this asymmetry in the structure of the DNA molecule, it is possible to
distinguish the two ends. The “top” of the strand is known as the 5′ end, and the
“bottom” as the 3′ end.
• A DNA sequence is read from the 5′ to the 3′ end.
• A DNA molecule comprises two strands of DNA which intertwine in a right-handed
“double helix” structure. The two strands are the exact complement of each other
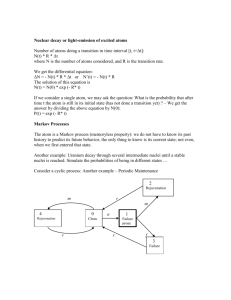
as A always pairs with T and G pairs with C; see Figure 4
Figure 4: Schematic of DNA structure
• Therefore, DNA can be studied by looking at only one of the strands, read in the
5′ to the 3′ direction.
58
MAS3326/8326 Discrete Stochastic Modelling
Notation
For the purposes of this part of the course we shall consider a DNA sequence of length n as
a string of letters y1 , y2 , . . . , yn from the alphabet S = {A, C, G, T}. The letters represent
the four nucleotides or bases.
Example 5.1
The first 1020 base pairs (bp) of the complete DNA sequence of bacteriophage lambda, a
parasite of the intestinal bacterium E-coli are given below.
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
GGGCGGCGAC
TTCTTCTTCG
ACAGGTGCTG
GGAATGAACA
TACCATTCAG
TGAGGTGCTT
TGAGAACGAA
CCAGCCAGGA
GGAACTGAAG
GCTGTCGCGG
GCGGCGTTTT
AGCCATGAAC
CGAACAGTCA
GGCCGGAGCC
CCGCATACCA
TGGGCAGCGA
AAATGCTGCT
CTCGCGGGTT
TCATAACTTA
AAAGCGAGGC
ATGGAAGTCA
AACTGGCAGG
TATGACTCTG
AAGCTGCGCC
ACTATTGAGT
AATGCCAGAG
ATCGCAGGTG
CCGGAACTGG
AAAGCAGCCG
GGTTAACAGG
ACAGACCGCC
GGAAGGGCGC
CTACATCCGT
GGGTGTTTAT
TTCGCTATTT
ATGTTTTTAT
TTTTTGGCCT
ACAAAAAGCA
AACAGGGAAT
CCGCCGTCAT
GGGAGGTTGA
ACGAACGCCA
ACTCCGCTGA
AAATTGCCAG
AAAACCGACA
CGCTGGATGA
CTGCGGCATT
GTTGAATGGG
TGGGAAACAC
GAGGTGAATG
GCCTACTTTA
ATGAAAATTT
TTAAAATACC
CTGTCGTTTC
GCTGGCTGAC
GCCCGTTCTG
AAAATGGTAT
AGAACTGCGG
TCGACTTACG
AGTGGTGGAA
TATTCTCGAC
TGTTGATTTC
ACTGATACCG
TTGTCCGCGC
CGGATGCTAA
TGCCCTTTCA
TGGTGAAGTC
TAGAGCATAA
TCCGGTTTAA
CTCTGAAAAG
CTTTCTCTGT
ATTTTCGGTG
CGAGGCGGTG
GCCGAAAGGG
CAGGCCAGCG
CGTGCGCAGG
ACCGCATTCT
GGGCTCCCCC
CTGAAACGGG
GGGTTGCTGA
CGGGCTTCGC
TTACTATCTC
GCGGGCCATC
TGCCCGTGTC
GCAGCGCAAC
GGCGTTTCCG
AAAGGAAACG
TTTTGTCCGT
CGAGTATCCG
GCAAGGGTAA
ATGCTGAAAT
AGGCAGATCT
CCGACGCACA
GTACTTTCGT
TGTCGGTGCA
ATATCATCAA
GTGAATATAT
TCACTGTTCA
CCGAAAGAAT
ATGAATGCGA
GGTTATTCCA
ACCCTTATCT
Therefore y1 = G, y2 = G, y3 = G, y4 = C, and so on.
59
MAS3326/8326 Discrete Stochastic Modelling
Motivation for modelling DNA sequences
Probabilistic/statistical models for DNA sequences have proved to be very useful and
important in real-world applications. For example, a particular class of models called
hidden Markov models (HMMs) have been of fundamental importance in gene finding
applications, and in determining regions of different DNA composition within the large
stretches of so-called “junk DNA” which, as yet, have no known function. Simpler models
such as those based on Markov chains assume that the DNA sequence is homogeneous,
that is, it has the same composition throughout. These models have been used to provide
a background model for determining whether some “words” (short strings of DNA) are
over- or under-represented within a DNA sequence of unknown function. In this part of
the module we will look at such models, but we begin by looking at perhaps the simplest
possible model for a DNA sequence, the independence model.
5.2
Independence model
Notation
Suppose that the DNA sequence y1 , y2, . . . , yn is a realisation of the random variables
Y1 , Y2 , . . . , Yn , where Yt is the base at location or site t in the sequence. Each Yt is a
categorical random variable with state space S = {A, C, G, T}.
The most simple model worth considering is the independence model which assumes
that the Yt are independent random variables and that, for t = 1, 2, . . . , n
Pr(Yt = A) = pA ,
Pr(Yt = C) = pC ,
Pr(Yt = G) = pG ,
Pr(Yt = T) = pT .
Here, pA , pC , pG , pT are called the base probabilities or emission probabilities. Clearly,
pA + pC + pG + pT = 1 and pi ≥ 0 for i ∈ S.
Another way of thinking about the independence assumption is to see that it imposes the
same base probabilities at a given site regardless of what bases preceded it, that is
Pr(Yt
Pr(Yt
Pr(Yt
Pr(Yt
=A
=C
=G
=T
|Yt−1, Yt−2, Yt−3, . . . , Y1) = Pr(Yt = A) = pA
|Yt−1, Yt−2, Yt−3, . . . , Y1) = Pr(Yt = C) = pC
|Yt−1, Yt−2, Yt−3, . . . , Y1) = Pr(Yt = G) = pG
|Yt−1, Yt−2, Yt−3, . . . , Y1) = Pr(Yt = T) = pT.
60
MAS3326/8326 Discrete Stochastic Modelling
Parameter estimation
It is straightforward to fit this model to any particular sequence. This can be done using
maximum likelihood to obtain estimates for the base probabilities p = (pA , pC , pG , pT )T .
Example 5.2 - constructing the likelihood
The likelihood function for p based on the sequence y = (A, C, G, T, A)T
is
L(p | y) = Pr(Y1 = A, Y2 = C, Y3 = G, Y4 = T, Y5 = A|p)
= Pr(Y1 = A) Pr(Y2 = C|Y1 = A) Pr(Y3 = G|Y2 = C, Y1 = A)×
× Pr(Y4 = T|Y3 = G, Y2 = C, Y1 = A)×
× Pr(Y5 = A|Y4 = T, Y3 = G, Y2 = C, Y1 = A)
= Pr(Y1 = A) Pr(Y2 = C) Pr(Y3 = G) Pr(Y4 = T) Pr(Y5 = A)
= pA pC pG pT pA
= p2A pC pG pT.
More generally, for a sequence y = (y1 , y2 , . . . , yn )T in which A occurs nA times, C nC times,
G nG times and T nT times, the likelihood function is
Y
pni i .
L(p | y) = pnA A pnC C pnG G pnT T =
i∈S
Maximum likelihood estimates can be found by maximising this function. One slight
complication is that there is not a free choice of values for the base probabilities as they
must sum to one.
Result 5.1
Given a DNA sequence y = (y1 , y2, . . . , yn )T in which A occurs nA times, C nC times, G nG
times and T nT times, then assuming an independence model the maximum likelihood
estimate (m.l.e.) of the base probability pi is
pbi =
ni
n
for i ∈ S. In other words, the m.l.e. is given by the sample proportion.
61
MAS3326/8326 Discrete Stochastic Modelling
Derivation of Result 5.1
The likelihood function for p based on data y is
L(p|y) =
Y
pni i .
i∈S
Therefore the loglikelihood is
ℓ(p|y) =
X
ni log pi.
i∈S
Since the elements of p are constrained to sum to 1 we wish to
P
maximise ℓ(p|y) subject to the constraint i∈S pi = 1. Therefore
introduce Lagrange multiplier λ and Lagrangian
!
X
X
L=
ni log pi − λ
pi − 1 .
i∈S
i∈S
Differentiating with respect to pi and setting equal to 0, gives
∂L ni
= − λ = 0.
∂pi pi
This stationary point of the loglikelihood function is a maximum
since
∂ 2L
ni
=
−
≤ 0.
∂p2i
p2i
This implies that
pi = ni/λ,
P
but i∈S pi = 1, therefore
P
i∈S ni
= 1,
λ
62
MAS3326/8326 Discrete Stochastic Modelling
P
i.e. λ = i∈S ni = n. Therefore the maximum likelihood estimate
of pi is given by
ni
pbi =
n
as required.
Example 5.3
Compute the maximum likelihood estimates of pA , pC , pG , pT based on the first 50 bases
of the bacteriophage lambda genome (see Example 5.1):
GGGCGGCGAC CTCGCGGGTT TTCGCTATTT ATGAAAATTT TCCGGTTTAA
Solution
The “counts” of each of the 4 letters are nA = 9, nC = 10, nG = 14
and nT = 17. Therefore the maximum likelihood estimates of the
base probabilities are
nA
n
nC
pbC =
n
nG
pbG =
n
nT
pbT =
n
pbA =
9
50
10
=
50
14
=
50
17
=
50
=
= 0.18
= 0.20
= 0.28
= 0.34.
Therefore maximum likelihood estimates the base probabilities by their sample equivalents, the observed proportion of As, Cs, Gs and Ts. Further, standard statistical theory
tells us the distributions that these estimates follow. For example, nA ∼ Bin(n, pA ) and
so, for large n, we have
pA (1 − pA )
pbA ∼ N pA ,
,
n
63
MAS3326/8326 Discrete Stochastic Modelling
with similar results for pbC , pbG and pbT . Therefore, for long sequences, (approximate) 95%
confidence intervals can be calculated easily. For example, for pA we use
!
r
r
pbA (1 − pbA )
pbA (1 − pbA )
.
pbA − 1.96 ×
, pbA + 1.96 ×
n
n
Example 5.4
Compute an approximate 95% confidence interval for pT based on the first 50 bases of the
bacteriophage lambda genome.
Solution
From Example 5.3, we have pbT = 0.34, and n = 50. Therefore an
approximate 95% confidence interval for pT is
!
r
r
0.34(1 − 0.34)
0.34(1 − 0.34)
0.34 − 1.96 ×
, 0.34 + 1.96 ×
,
50
50
that is
(0.2087,
0.4713).
The independence model provides a very basic model for DNA sequences. It can be
used to determine whether various “words” are over- or under-represented relative to
the “background” probability. For example, the independence assumption implies that
the number of sites between occurrences of words has a Geometric distribution with an
appropriate probability parameter.
However, there is evidence to suggest that DNA sequences are not independent strings of
letters and so more sophisticated models may be more appropriate.
64
MAS3326/8326 Discrete Stochastic Modelling
5.3
5.3.1
Markov chain models
Introduction
A major drawback with the very basic model described in the previous section is that the
independence assumption does not capture the complex base dependencies known to exist
in DNA sequences. However, the model can be generalised to allow base probabilities to
depend on the bases at previous sites by using Markov chain models.
Reminder of Definition 2.1
The random variables Y1 , Y2, . . . form a Markov chain with state space S if
Pr(Yt+1 = j|Yt = i, Yt−1 , . . . , Y1 ) = Pr(Yt+1 = j|Yt = i) = pij (t)
for all t and for i, j ∈ S.
The Markov chain is said to be homogeneous if pij (t) = pij for all t.
5.3.2
Markov chain models for DNA sequences
In terms of a DNA sequence, in a homogeneous Markov chain model, the base probabilities
are allowed to depend on the base at the previous site, for example
Pr(Yt = C|Yt−1 = A, Yt−2 , Yt−3 , . . . , Y1 ) = Pr(Yt = C|Yt−1 = A) = pAC .
Similar transition probabilities are used to describe other changes between adjacent
sites and are summarised by the model’s transition matrix
pAA pAC pAG pAT
pCA pCC pCG pCT
P =
pGA pGC pGG pGT .
pTA pTC pTG pTT
The top row describes how, as we move along the sequence, base A is followed by each
of the four possibilities A, C, G, T. Recall that the probabilities along each row must sum
to one as some transition has to occur. Matrices with this property are used often in the
study of discrete processes which evolve discretely in time and we recall that these are
termed stochastic matrices.
The Markov chain model described above is often called a first order Markov chain
model, as the probability of the base at site t depends only on the first base preceding it.
The first order Markov model is a generalisation of the independence model as this model
65
MAS3326/8326 Discrete Stochastic Modelling
can be obtained as a special case. This occurs when the entries are the same within the
column, that is
pA pC pG pT
pA pC pG pT
P =
pA pC pG pT .
pA pC pG pT
Therefore, the independence model can be thought of as a Markov chain model in which
transition probabilities depend on the previous zero bases, that is, a zero order Markov
chain model.
5.3.3
Parameter inference
Likelihood function
Returning to the first order Markov chain model, the likelihood function for the transition
probabilities P is
L(P |y) = Pr(Y1, Y2, . . . , Yn)
= Pr(Y1) Pr(Y2|Y1) Pr(Y3|Y1, Y2) · · · Pr(Yn|Y1, Y2, . . . , Yn−1 )
= Pr(Y1) Pr(Y2|Y1) Pr(Y3|Y2) · · · Pr(Yn|Yn−1)
= Pr(Y1) × py1y2 py2y3 · · · pyn−1yn
n−1
Y
= Pr(Y1) ×
pytyt+1
Yt=1 nij
= Pr(Y1)
pij ,
i,j∈S
where
nij =
n−1
X
I(yt+1 = j|yt = i)
t=1
is the number of times base i is followed by base j. Hence, nij represents the number of
occurrences in the sequence of an i → j transition. (A pair of consecutive bases, e.g. AG,
is called a dinucleotide.)
66
MAS3326/8326 Discrete Stochastic Modelling
Dealing with the initial probability
The likelihood function is complicated by the first term Pr(Y1 ). As we have a homogeneous
sequence in which the transition probabilities are the same at every point in the sequence,
we could assume that
Pr(Y1 = A) = Pr(Y2 = A) = · · · = Pr(Yn = A) = Pr(A).
This should be interpreted as saying that the probability of an A at any particular location
is the same throughout the sequence if we do not know the base in the previous position.
Of course, if we did know the previous base was, say a T then this probability changes
to pTA . These marginal probabilities are called stationary probabilities and we have
met them in Section 2! Recall that these probabilities have a special (row-vector) notation
π = (πA , πC , πG , πT ). They are calculated using the Law of Total Probability. For example,
πA = Pr(A) = Pr(A|A) Pr(A) + Pr(A|C) Pr(C) + Pr(A|G) Pr(G) + Pr(A|T) Pr(T)
= pAA πA + pCA πC + pGA πG + pTA πT .
Similar expressions are available for the other stationary probabilities. This gives a system
of equations in four unknowns which can be summarised as
X
πj =
πi pij
i∈S
or, in matrix notation, as
π = πP.
Recall that due to the special structure of stochastic matrices, this system contains only
three “independent” equations. Therefore an additional condition must be used to determine π, namely that the stationary probabilities must sum to one:
X
πi = 1.
i∈S
The vector π represents the stationary distribution of the Markov chain.
Returning to the likelihood function, the inclusion of Pr(Y1 ) presents some difficulties
due to its complex dependence on the transition probability parameters. One solution is
to consider an additional component to the model that describes the initial base in the
sequence, for example, we might assume that all four bases are equally likely (Pr(Y1 ) =
1/4). An alternative argument asserts that the information in this first base will be
dominated by that in the rest of the sequence and so very little will be lost if this term is
ignored (and generally we’ll adopt this approach in this course).
Both arguments lead to using the likelihood function
Y n
L(P |y) ∝
pijij .
i,j∈S
67
MAS3326/8326 Discrete Stochastic Modelling
Typically, in a first order Markov chain model we will consider inference conditional on
the first base in the sequence, hence the likelihood function is
Y n
L(P |y) =
pijij .
i,j∈S
Maximum likelihood estimation
We now have the likelihood function for the transition probabilities. The next step is to
obtain maximum likelihood estimates for the transition probabilities by maximising the
likelihood function.
Result 5.2
Given a DNA sequence y = (y1 , y2 , . . . , yn )T and assuming a homogeneous first order
Markov chain model, the maximum likelihood estimate of the transition probability
pij (conditional on the first observation y1 ) is
nij
pbij = P
j∈S nij
for i, j ∈ S, where nij denotes the number of occurrences in the sequence of an i → j
transition.
Derivation of Result 5.2
Let P = (pij ) be the matrix of transition probabilities, i, j ∈ S. The likelihood function
for P based on data y (and conditional on y1 ) is
L(P |y) =
Y
n
pijij ,
i,j∈S
and therefore the loglikelihood is
X
nij log pij .
ℓ(P |y) =
i,j∈S
Since the rows of P are constrained to sum to 1 we wish to maxP
imise ℓ(P |y) subject to the constraint j∈S pij = 1. Therefore
introduce Lagrange multiplier λ and Lagrangian
X
X
L = constant +
nij log pij − λ
pij − 1 .
i,j∈S
j∈S
68
MAS3326/8326 Discrete Stochastic Modelling
Differentiating with respect to pij and setting equal to 0, gives
∂L
nij
=
− λ = 0,
∂pij
pij
and this is a maximum since
∂ 2L
nij
=
−
≤ 0.
∂p2ij
p2ij
This implies that
but
P
j∈S
pij = nij /λ,
pij = 1, therefore
P
j∈S
nij
λ
= 1,
P
i.e. λ = j∈S nij .
Therefore the maximum likelihood estimate is given by
nij
pbij = P
j∈S nij
as required.
69
MAS3326/8326 Discrete Stochastic Modelling
Example 5.5
The values of nij (i, j ∈ S) obtained from the first 1020 bp of the bacteriophage lambda
genome are given below.
j
i
A
C
G
T
A
86
47
58
58
C
54
55
72
59
G
73
84
77
53
T
36
54
79
74
Compute the maximum likelihood estimates of the transition probabilities pij based on
these data. Report the estimates to three decimal places.
Solution
P
P
P
We have,
n
=
249,
n
=
240,
j Aj
j Cj
j nGj = 286 and
P
j nTj = 244, therefore,
nAA
86
≃ 0.345,
pbAA = P
=
249
n
j Aj
and so on. This gives m.l.e.s for the transition probabilities (rounded
to 3dp) as
0.345 0.217 0.293 0.145
0.196
0.229
0.350
0.225
Pb =
.
0.203 0.251 0.269 0.276
0.238 0.242 0.217 0.303
Just as with the independence model of Section 5.2, standard statistical theory can be
used to determine (approximate) confidence intervals for the transition probabilities pij .
70
MAS3326/8326 Discrete Stochastic Modelling
5.4
Model choice
In Example 5.5 we fitted a first order Markov chain model to the first 1020 bases of the
bacteriophage lambda sequence. We could also have fitted an independence model to this
sequence (i.e. a zero order Markov chain). But which is better? The zero order model is
simpler than the first order model as it has fewer parameters and so is easier to interpret,
but the more complex first order model will provide a better fit to the data (in terms of
the likelihood).
When choosing between Markov chain models we generally adopt the principle of parsimony and favour a simpler model over a more complex model provided the fit to the
data is similar.
The Schwarz criterion1 is a commonly used method for model choice that provides
a trade-off between model complexity (as measured by the number of parameters) and
model fit (as measured by the loglikelihood evaluated at the maximum likelihood estimates
of the parameters).
Definition 5.1 – Schwarz criterion
Let k denote the number of free parameters in a model and let θb denote the maximum
likelihood estimates of the parameters θ from the model. The value of the loglikelihood
b The Schwarz criterion is defined as
evaluated at the MLE is denoted ℓ(θ).
b + k log(m),
S = −2ℓ(θ)
where m is the number of datapoints used to fit the model.
Remarks
• The Schwarz criterion is calculated for all competing models and the model with
the smallest value of the Schwarz criterion is the favoured model.
• For Markov chain models, as the order of dependence increases the value of the
b also increases, indicating better fit to the data.
maximised loglikelihood ℓ(θ)
• However, the number of free parameters also increases with increased order and this
is penalized in the Schwarz criterion.
• Therefore the “best” model is not necessarily the model with the most parameters.
• It can be shown that the Schwarz criterion leads to consistent estimation of the
order of a Markov chain.
1
Also known as the Bayesian Information Criterion (BIC)
71
MAS3326/8326 Discrete Stochastic Modelling
Example 5.6
Consider again the first 1020 bp of the bacteriophage lambda genome that we looked at
in Example 5.1. The number of i → j transitions nij (i, j ∈ S) are given below.
j
i
A
C
G
T
A
86
47
58
58
C
54
55
72
59
G
73
84
77
53
T
36
54
79
74
Now suppose we wish to choose between a first order Markov chain model and a zero
order Markov chain model for this DNA sequence. For each model we must
(i) compute the maximum likelihood estimates θb of the parameters in the model;
b
(ii) compute the maximised loglikelihood ℓ(θ);
(iii) compute the number of free parameters in the model k;
b + k log(m), where m is the number of
(iv) compute the Schwarz criterion, S = −2ℓ(θ)
datapoints used to fit the model.
We then choose the model with the smallest value of the Schwarz criterion.
Consider first the first order Markov model that we looked at in Example 5.5.
(i) The MLEs of the transition probabilities P
given below:
0.345 0.217
0.196 0.229
Pb =
0.203 0.251
0.238 0.242
were computed in Example 5.5 and are
0.293
0.350
0.269
0.217
0.145
0.225
.
0.276
0.303
(ii) The formula for the loglikelihood conditional on the first base y1 (see Derivation of
Result 5.2) is
X
ℓ(P ) =
nij log pij .
i,j∈S
Therefore the maximised loglikelihood is
72
MAS3326/8326 Discrete Stochastic Modelling
ℓ(Pb) =
X
i,j∈S
nij log pbij
= 86 × log(0.345) + 54 × log(0.217) + · · · + 74 × log(0.303)
= −1390.387.
(iii) There are 4 × 4 = 16 transition probabilities, therefore 16 parameters in the first
order Markov chain model. However, we do not have a free choice over the values
of all these parameters; the row sums are constrained to be equal to 1. Therefore
we only have 4 × 3 = 12 free parameters, so k = 12.
(iv) In order to compute the value of the Schwarz criterion S, we also need the number
of datapoints m that were used to fit the model. Although we have a sequence of
length n = 1020, we conditioned on the first base y1 when fitting the model, so we
have used m = 1019 datapoints. Alternatively,
Pwe can compute m by summing up
the number of i → j transitions, that is m = i,j∈S nij .
Therefore,
b + k log(m)
S = −2ℓ(θ)
= −2 × −1390.387 + 12 × log(1019)
= 2780.774 + 83.119
= 2863.893.
We now consider the zero order Markov model (i.e. the independence model) and follow
the same four-step procedure. Crucially, we must fit the model to the same data that
was used to fit the first order model. This means estimating parameters conditional on
the first base y1 and thus using only m = 1019 datapoints rather than n = 1020. It also
means that we must use the transition counts (the nij ) to estimate the base probabilities.
(i) Recall from Result 5.1 that the MLE of the base probability pi for i ∈ S is
pbi =
73
ni
n
MAS3326/8326 Discrete Stochastic Modelling
where ni represents the number of occurrences of the base i in the sequence and n
represents the number of bases in the whole sequence. Since we are now conditioning
on the first base in the sequence the corresponding expression for the MLE can be
shown to be
P
P
n
ji
j∈S
j∈S nji
pbi = P
.
=
m
i,j∈S nij
In other words, this is simply the total number of transitions to base i from any
other base divided by the total number of transitions m.
So, for the bacteriophage lambda sequence we have
pbA =
249
≃ 0.244,
1019
pbC =
240
≃ 0.236,
1019
pbG =
287
≃ 0.282,
1019
pbT =
243
≃ 0.238.
1019
(ii) The formula for the loglikelihood conditional on the first base y1 is
X X
ℓ(p) =
nji log pi.
i∈S
j∈S
Therefore the maximised loglikelihood is
X X
nji log pbi
ℓ(b
p) =
i∈S
j∈S
= 249 × log(0.244) + 240 × log(0.236) + 287 × log(0.282) + 243 × l
= −1409.899.
Note that the maximised loglikelihood under this zero order model is smaller than
the maximised loglikelihood under the more complex first order model.
(iii) There are 4 base probabilities, but they are restricted to sum to one, meaning that
there are only k = 3 free parameters in the zero order model.
(iv) We have fitted the zero order model using the same number of datapoints that we
used to fit the first order model, that is m = 1019, so the value of the Schwarz
criterion is
74
MAS3326/8326 Discrete Stochastic Modelling
b + k log(m)
S = −2ℓ(θ)
= −2 × −1409.899 + 3 × log(1019)
= 2819.798 + 20.780
= 2840.587.
We then choose the model with the smaller value of the Schwarz
criterion, which in this case is the zero order model (2840.587 <
2863.893).
This means that, although the first order model provides a better fit to the data in terms
of the loglikelihood, this improved fit is accomplished at the expense of fitting many more
parameters which isn’t justified for this DNA sequence.
5.4.1
General qth order Markov chain models
A Markov chain model of order q (≥ 0) is loosely defined as a stochastic process in which
the probability of the current state depends on only the values of the previous q states,
that is
Pr(Yt |Yt−1 , Yt−2 , . . . , Y1 ) = Pr(Yt |Yt−1 , Yt−2 , . . . , Yt−q ).
Such a qth order Markov chain model is denoted M(q). So, for example, M(0) denotes
the independence model, M(1) denotes the standard first order Markov chain model, and
so on.
Maximum likelihood parameter estimation
First condition on the first q bases in the sequence and then count the number of qth
order transitions i1 → i2 → · · · → iq → iq+1 (i1 , i2 , . . . , iq , iq+1 ∈ S) and denote these
ni1 i2 ...iq iq+1 . In other words
ni1 i2 ...iq iq+1 =
n
X
I(yt−q = i1 , yt−q+1 = i2 , . . . , yt−1 = iq , yt = iq+1 ).
t=q+1
75
MAS3326/8326 Discrete Stochastic Modelling
It can be shown that the maximum likelihood estimate of the qth order transition probability pi1 i2 ...iq iq+1 is
ni1 i2 ...iq iq+1
pbi1 i2 ...iq iq+1 = P
.
iq+1 ∈S ni1 i2 ...iq iq+1
The value of the maximised loglikelihood for an M(q) model is
X
ℓ(θbq ) =
ni1 i2 ...iq iq+1 log(b
pi1 i2 ...iq iq+1 ),
i1 ,i2 ,...,iq ,iq+1 ∈S
where θbq denotes the maximum likelihood estimate of the parameters of the M(q) model.
The number of free parameters in an M(q) model with state space S is kq = bq (b − 1)
where b = |S| denotes the number of states (which for DNA sequences is b = 4).
Model choice
Suppose we wish to choose the “best” Markov chain model for a DNA sequence y1 , y2 , . . . , yn .
We entertain models of order q = 0, 1, . . . , qmax where qmax ≥ 0. We first consider the
maximal model (i.e. the largest model with the most parameters) M(qmax ), and compute
the transition counts ni1 i2 ...iqmax +1 conditional on the first qmax observations. These transition counts are used to derive maximum likelihood estimates for all models. This means
that the number of datapoints used for all models is m = n − qmax . Then for each model
q = 0, 1, . . . , qmax , we compute the Schwarz criterion
Sq = −2ℓ(θbq ) + kq log(m).
Then, as before, we choose the model with the smallest value of Sq .
Example 5.7
In this example we’ll look at choosing between Markov models of order q = 0, 1 and 2 for
the DNA sequence of the gorilla mitochondrial genome (n = 16364 bp).
The number of each of the 64 possible 2nd order transition counts (the triplets i1 → i2 →
i3 ) are given below.
76
MAS3326/8326 Discrete Stochastic Modelling
i3
i1
A
A
A
A
C
C
C
C
G
G
G
G
T
T
T
T
i2
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
514
421
175
374
451
466
112
523
204
191
123
164
417
403
204
317
C
480
501
271
382
441
596
142
401
154
258
142
109
362
357
125
301
G
204
118
168
174
194
135
77
183
124
45
71
70
251
120
109
116
T
388
397
159
332
394
515
87
304
132
186
89
98
348
313
105
275
The table below gives the maximised loglikelihoods ℓ(θbq ) under each model.
0
1
2
q
b
ℓ(θq ) −21926.26 −21792.51 −21714.89
For each model, the number of datapoints used is
m = n − qmax
= 16364 − 2 = 16362.
Also, the number of free parameters in each model is
k0 = 40(4 − 1) = 1 × 3 = 3
k1 = 41(4 − 1) = 4 × 3 = 12
k2 = 42(4 − 1) = 16 × 3 = 48.
77
MAS3326/8326 Discrete Stochastic Modelling
Therefore the value of the Schwarz criterion for each model is
S0 = − 2 × −21926.26 + 3 × log(16362) = 43852.52 + 29.12 = 43881.64
S1 = − 2 × −21792.51 + 12 × log(16362) = 43585.02 + 116.43 = 43701.45
S2 = − 2 × −21714.89 + 48 × log(16362) = 43429.78 + 465.73 = 43895.51.
S1 is the smallest value, so the M (1) model is preferred for these
data.
Limitations of maximum likelihood
One potential drawback of using the maximum likelihood approach to inference for Markov
chain models is that for high order models there may not be sufficient data to accurately
estimate the transition probability parameters.
For example, suppose we have a relatively short DNA sequence, say n = 1000. If we try
to fit an M(4) model there are 44 (4 − 1) = 768 free transition probabilities to estimate.
The average number of transitions of a particular type is roughly 1000/768 ≃ 1.3. This
is small, and it is quite likely that we may not observe any 4th order transitions of a
particular type. The corresponding MLE of the transition probability would be 0.
Worse still, when considering even higher order models, we may encounter the situation
in which
X
ni1 i2 ...iq iq+1 = 0,
iq+1 ∈S
in which case the MLE is not defined.
These drawbacks may be easily overcome by using a Bayesian approach to inference
(details omitted!).
78