Determining Protein Concentration by Modified Micro
advertisement

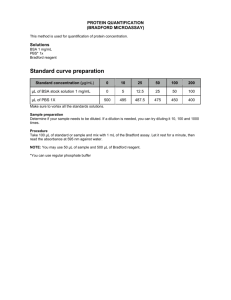
Determining Protein Concentration by Modified Micro-Bradford Assay The Modified Micro-Bradford Assay Note: This assay protocol is intended to quantify protein samples containing any reagents. It has been adapted from the Bio-Rad Bradford protein assay kit (#500-0006). For a list of directly compatible reagents see the BioRad Bradford assay technical manual (technical note 1069). This protocol only addresses issues of protein quantification. Based on the composition of your sample buffer (against the compatibility chart: Appendix A) chose whether to perform the standard or modified Bradford assay as outlined below. The Standard Bradford: This procedure is an adaptation of BioRad’s Bradford micro-assay – set up in a 96 well flat bottom plate. The standard curve requires 80µg of BSA or IgG, and you will need one standard curve for every 7 samples. Ideally, you should also assay approximately 80µg of each unknown sample in order to obtain the most data points, but this assay will be able to determine final protein concentration as long as 0.2-5 mg/mL. 1 Perform the protein assay in according to the standard Bradford methodologies above. 2 Select a volume of your unknown protein samples to assay. Make up to 80 µL with dH2O, or appropriate compatible buffer. **Here you have to ensure that the final concentration of SDS is <0.05% and Urea is <6M** 3 Take 80 ug of standard and also make up to 80 µL in an appropriate buffer. **Here an appropriate buffer is one that will make the final reagent concentrations (ex. SDS, Urea…ect.) the same as the unknown protein samples. For example, if you had a sample in 0.1% SDS, 6M urea, and you chose to assay 20 µL – diluting in 60 µL of dH2O – your final concentrations are 0.025% SDS and 1.5M urea.If your standard was BSA at 2 mg/mL in dH2O or weak buffer, you would need to add 40 µL of standard to 40 µL of 0.05% SDS and 3M Urea, so your final solution would be 80 µg of BSA in 80 µL of 0.025% and 1.5M urea. 4 You also need to make a dilution buffer with components equivalent to your unknown protein samples and standard ~ 1 mL per 96 well plate. If your samples were prepared according to the above example, your dilution buffer would also be 0.025% SDS and 1.5M Urea. Column: 1 2 4 5 6 7 8 9 10 11 12 Sample 10 µL 10 µL 8 µL 8 µL 6 µL 6 µL 4 µL 4 µL 2 µL 2 µL 0 µL 0 µL Dilution 0 µL 2 µL 4 µL 4 µL 6 µL 6 µL 8 µL 8 µL 10 µL 10 µL 0 µL 3 2 µL This gives you 5 different dilutions of each sample and a standard curve from 0-10 µg per well, all in duplicate. 5 Set-up each row of the 96-well plate as follows, using one row per standard/sample: 6 To develop assay add 200 µL of 1:5 diluted Bradford reagent (BioRad 500-0006) to each well, mix, and allow 5-10 min at room temperature for development. 7 Within 60 min of development read the plate at 595nm. The Modified Bradford: This procedure is intended for protein preparations containing reagents at concentrations that are incompatible with the Bradford reagent. However, all samples – including the 80 µg of BSA or IgG used for the standard curve - need to be precipitated and resolubilized in a compatible buffer before the assay. a Protein precipitation. Protein Precipitation Protocols Acetone/ TCA Precipitation ( works best for dilute samples) 1 Mix 10 volumes of cold 10% TCA in acetone (stored –20C) with your samples, vortex, and incubate at –20C for at least 3hrs, but overnight is optimal. 2 Centrifuge samples at 15000g X 10min and remove supernatant. 3 Add the same volume of cold acetone only, vortex, and let stand at – 20C for at least 10min. 4 Centrifuge at 15000g X 5min, remove supernatant and allow pellets to air dry. Care should be taken to prevent complete desiccation of the protein pellet as this will make re-solubilization much harder. TCA / DOC Precipitation Notes: DOC is a detergent and must be removed from the sample solution prior to analysis by MS. 1 Mix your sample with 1/100 of its volume of 2% DOC and incubate on ice for 30 minutes. 2 Add enough 100% TCA to bring your sample to a final TCA concentration of 15% and votex immediately for 30 seconds to prevent any large conglomerates from forming that may trap the contaminants you are trying to remove. 3 Let the sample precipitate for a minimum of one hour, but preferably overnight. 4 Spin out the precipitate at 15,000g for 10 minutes and aspirate the TCA and soluble contaminants from the pellet. 5 Wash the pellet with ice cold ethanol or acetone making sure to break up the pellet to the point that it is getting washed. 6 Vortex and let sit at room temp for 5 minutes. 7 Spin down the pellet at 15,000g for 10 minutes and aspirate the supernatant off of the pellet. 8 Repeat the wash step and re-spin down the pellet. 9 Dry the pellet under a SLOW stream of nitrogen and make sure not to bring the pellet to complete dryness as it will be difficult to bring back into solution. Chloroform / Methanol Precipitation 1 Add 4 volumes of methanol and vortex well. 2 Add 1 volume of chloroform and vortex. 3 Add 3 volumes of dH2O and vortex. 4 Centrifuge for 2min at 15 000g – the proteins should be at the liquid interface. 5 Remove the aqueous top layer and add 4 volumes of methanol – vortex. 6 Centrifuge for 2min at 15 000g. 7 Remove as much liquid as possible without disturbing precipitate. 8 Speed-Vac sample to dryness or dry under nitrogen. Protein Re-solubilization The protein precipitates need to be re-solubilized in a buffer compatible with the next step of analysis. For example: SDS-PAGE sample buffer for 1D gel analysis, re-hydration buffer for 2D-gel analysis, or a small volume of fresh buffered 6M urea then diluted to an appropriate concentration for an in-solution digests and protein quantification. b Protein Resolubilization: • • • • Add 80 µL of resolubilization buffer to each protein precipitate. Resolubilization buffer – 0.05% SDS and 6M Urea. The sample can be quickly heated to help solvation. Centrifuge 15000g X 5min and check for a pellet to ensure full protein resuspension. Perform the protein assay in according to the standard Bradford methodologies above.