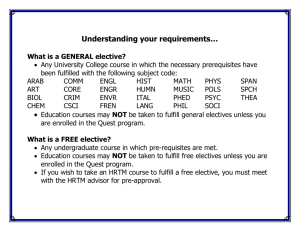
Essential Cell Biology, Fourth Edition
advertisement

Essential Cell Biology, Fourth Edition REFERENCES Chapter 1 The Unity and Diversity of Life Andersson SGE (2006) The bacterial world gets smaller. Science 314:259–260. Brenner S, Jacob F & Meselson M (1961) An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190:576– 581. Fraser CM, Gocayne JD, White O et al (1995) The minimal gene complement of Mycoplasma genitalium. Science 270:397–403. Harris JK, Kelley ST, Spiegelman et al (2003) The genetic core of the universal ancestor. Genome Res 13:407–413. Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39:309–338. Watson JD & Crick FHC (1953) Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 171:737–738. Yusupov MM,Yusupova GZ, Baucom A et al (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883–896. Rhinn M et al. (2009) Zebrafish gbx1 refines the midbrain-­‐hindbrain boundary border and mediates the Wnt8 posteriorization signal. Neural Dev 4:12. Looking at Cells in the Light Microscope Adams MC, Salmon WC, Gupton SL et al (2003) A high-­‐ speed multispectral spinning-­‐disk confocal microscope system for fluorescent speckle microscopy of living cells. Methods 29:29–41. Agard DA, Hiraoka Y, Shaw P & Sedat JW (1989) Fluorescence microscopy in three dimensions. In Methods in Cell Biology, vol 30: Fluorescence Microscopy of Living Cells in Culture, part B (DL Taylor, Y-­‐L Wang eds). San Diego: Academic Press. Centonze VE (2002) Introduction to multiphoton excitation imaging for the biological sciences. Methods Cell Biol 70:129–48. Chalfie M, Tu Y, Euskirchen G et al (1994) Green fluorescent protein as a marker for gene expression. Science 263:802–805. Egner A et al. (2002) Fast 100-­‐nm resolution three-­‐ dimensinoal microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci USA 99(6): 3370-­‐5. Giepmans BN, Adams SR, Ellisman MH & Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science 312:217–224. Harlow E & Lane D (1988) Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. Haugland RP (ed) (1996) Handbook of Fluorescent Probes and Research Chemicals, 8th ed. Eugene, OR: Molecular Probes, Inc. (Available online at http://www.probes.com/) Jaiswai JK & Simon SM (2004) Potentials and pitfalls of fluorescent quantum dots for biological imaging. Trends Cell Biol 14:497–504. Jares-­‐Erijman EA & Jovin TM (2003) FRET imaging. Nature Biotech 21:1387–1395. Lippincott-­‐Shwartz J, Altan-­‐Bonnet N & Patterson G (2003) Photobleaching and photoactivation: following protein dynamics in living cells. Nature Cell Biol 5:S7–S14. Michalet X et al. (2003) The power and prospects of fluorescence microscopies and spectroscopies. Annu Rev Biophys Biomol Struct 32:161-­‐82. Minsky M (1988) Memoir on inventing the confocal scanning microscope. Scanning 10:128–138. Miyawaki A, Sawano A & Kogure T (2003) Lighting up cells: labeling proteins with fluorophores. Nature Cell Biol 5:S1–S7. Parton RM & Read ND (1999) Calcium and pH imaging in living cells. In Light Microscopy in Biology. A Practical Approach, 2nd ed. (Lacey AJ ed) Oxford: Oxford University Press. Sako Y & Yanagida T (2003) Single-­‐molecule visualization in cell biology. Nature Rev Mol Cell Biol 4:SS1–SS5. Sekar RB & Periasamy A (2003) Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol 160:629–633. Shaner NC, Steinbach PA & Tsien RY (2005) A guide to choosing fluorescent proteins. Nature Methods 2:905–909. Sheetz MP (ed) (1997) Laser Tweezers in Cell Biology. Methods Cell Biol 55. Sluder G & Wolf DE (2007) Video Microscopy 3rd ed. Methods Cell Biol 81. Stevens DJ & Allan (2003) Light Microscopy Techniques for Live Cell Imaging. Science 300:82–86. Tsien RY (2003) Imagining imaging’s future. Nature Rev Mol Cell Rev 4:SS16–SS21. van Teeffelen S, Shaevitz JW, Gitai Z (2012) Image analysis in fluorescence microscopy: bacterial dynamics as a case study. Bioessays 34(5): 427-­‐36 Weiss DG, Maile W, Wick RA & Steffen W (1999) Video microscopy. In Light Microscopy in Biology: A Practical Approach, 2nd ed. (AJ Lacey ed) Oxford: Oxford University Press. Essential Cell Biology, Fourth Edition REFERENCES White JG, Amos WB & Fordham M (1987) An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J Cell Biol 105:41–48. Zernike F (1955) How I discovered phase contrast. Science 121:345–349. Looking at Cells and Molecules in the Electron Microscope Allen TD & Goldberg MW (1993) High resolution SEM in cell biology. Trends Cell Biol 3:203–208. Baumeister W (2002) Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol 12:679–684. Böttcher B, Wynne SA & Crowther RA (1997) Determination of the fold of the core protein of hepatits B virus by electron cryomicroscopy. Nature 386:88–91. Dubochet J, Adrian M, Chang J-­‐J et al (1988) Cryoelectron microscopy of vitrified specimens. Q Rev Biophys 21:129–228. Frank J (2003) Electron microscopy of functional ribosome complexes. Biopolymers 68:223–233. Hayat MA (2000) Principles and Techniques of Electron Microscopy, 4th ed. Cambridge: Cambridge University Press. Heuser J (1981) Quick-­‐freeze, deep-­‐etch preparation of samples for 3D electron microscopy. Trends Biochem Sci 6:64–68. Lippincott-­‐Schwartz J & Patterson GH (2003) Development and use of fluorescent protein markers in living cells. Science 300:87–91. McIntosh R, Nicastro D & Mastronarde D (2005) New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol 15:43–51. McDonald KL & Auer M (2006) High pressure freezing, cellular tomography, and structural cell biology. Biotechniques 41:137–139. Pease DC & Porter KR (1981) Electron microscopy and ultramicrotomy. J Cell Biol 91:287s–292s. Unwin PNT & Henderson R (1975) Molecular structure determination by electron microscopy of unstained crystal specimens. J Mol Biol 94:425–440. Chapter 2 The Chemical Components of a Cell Abeles RH, Frey PA & Jencks WP (1992) Biochemistry. Boston: Jones & Bartlett. Atkins PW (1996) Molecules. New York: WH Freeman. Branden C & Tooze J (1999) Introduction to Protein Structure, 2nd ed. New York: Garland Science. Bretscher MS (1985) The molecules of the cell membrane. Sci Am 253:100–109. Burley SK & Petsko GA (1988) Weakly polar interactions in proteins. Adv Protein Chem 39:125– 189. De Duve C (2005) Singularities: Landmarks on the Pathways of Life. Cambridge: Cambridge University Press. Dowhan W (1997) Molecular basis for membrane phospholipid diversity: Why are there so many lipids? Annu Rev Biochem 66:199–232. Eisenberg D & Kauzman W (1969) The Structure and Properties of Water. Oxford: Oxford University Press. Fersht AR (1987) The hydrogen bond in molecular recognition. Trends Biochem Sci 12:301–304. Franks F (1993) Water. Cambridge: Royal Society of Chemistry. Henderson LJ (1927) The Fitness of the Environment, 1958 ed. Boston: Beacon. Neidhardt FC, Ingraham JL & Schaechter M (1990) Physiology of the Bacterial Cell: A Molecular Approach. Sunderland, MA: Sinauer. Pauling L (1960) The Nature of the Chemical Bond, 3rd ed. Ithaca, NY: Cornell University Press. Saenger W (1984) Principles of Nucleic Acid Structure. New York: Springer. Sharon N (1980) Carbohydrates. Sci Am 243:90–116. Stillinger FH (1980) Water revisited. Science 209:451– 457. Tanford C (1978) The hydrophobic effect and the organization of living matter. Science 200:1012– 1018. Tanford C (1980) The Hydrophobic Effect. Formation of Micelles and Biological Membranes, 2nd ed. New York: John Wiley. White SH (2005) How hydrogen bonds shape membrane protein structure. Adv Protein Chem 72:157-­‐72. Chapter 3 Catalysis and the Use of Energy by Cells Atkins PW (1994) The Second Law: Energy, Chaos and Form. New York: Scientific American Books. Atkins PW & De Paula JD (2006) Physical Chemistry for the Life Sciences. Oxford: Oxford University Press. Baldwin JE & Krebs H (1981) The Evolution of Metabolic Cycles. Nature 291:381–382. Berg HC (1983) Random Walks in Biology. Princeton, NJ: Princeton University Press. Dickerson RE (1969) Molecular Thermodynamics. Menlo Park, CA: Benjamin Cummings. Essential Cell Biology, Fourth Edition REFERENCES Dill KA & Bromberg S (2003) Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. New York: Garland Science. Dressler D & Potter H (1991) Discovering Enzymes. New York: Scientific American Library. Einstein A (1956) Investigations on the Theory of Brownian Movement. New York: Dover. Fruton JS (1999) Proteins, Enzymes, Genes: The Interplay of Chemistry and Biology. New Haven: Yale University Press. Goodsell DS (1991) Inside a living cell. Trends Biochem Sci 16:203–206. Karplus M & McCammon JA (1986) The dynamics of proteins. Sci Am 254:42–51. Karplus M & Petsko GA (1990) Molecular dynamics simulations in biology. Nature 347:631–639. Kauzmann W (1967) Thermodynamics and Statistics: with Applications to Gases. In Thermal Properties of Matter Vol 2. New York: WA Benjamin, Inc. Kornberg A (1989) For the Love of Enzymes. Cambridge, MA: Harvard University Press. Lavenda BH (1985) Brownian Motion. Sci Am 252:70– 85. Lawlor DW (2001) Photosynthesis, 3rd ed. Oxford: BIOS. Lehninger AL (1971) The Molecular Basis of Biological Energy Transformations, 2nd ed. Menlo Park, CA: Benjamin Cummings. Lipmann F (1941) Metabolic generation and utilization of phosphate bond energy. Adv Enzymol 1:99–162. Lipmann F (1971) Wanderings of a Biochemist. New York: Wiley. Nisbet EE & Sleep NH (2001) The habitat and nature of early life. Nature 409:1083–1091. Racker E (1980) From Pasteur to Mitchell: a hundred years of bioenergetics. Fed Proc 39:210–215. Schrödinger E (1944 & 1958) What is Life?: The Physical Aspect of the Living Cell and Mind and Matter, 1992 combined ed. Cambridge: Cambridge University Press. Shi Y (2013) Common folds and transport mechanisms of secondary active transporters. Ann Rev Biophys 42: 51-­‐72. van Holde KE, Johnson WC & Ho PS (2005) Principles of Physical Biochemistry, 2nd ed. Upper Saddle River, NJ: Prentice Hall. Walsh C (2001) Enabling the chemistry of life. Nature 409:226–231. Westheimer FH (1987) Why nature chose phosphates. Science 235:1173–1178. Youvan DC & Marrs BL (1987) Molecular mechanisms of photosynthesis. Sci Am 256:42–49. Chapter 4 The Shape and Structure of Proteins Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230. Bray D (2005) Flexible peptides and cytoplasmic gels. Genome Biol 6:106–109. Burkhard P, Stetefeld J & Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11:82–88. Caspar DLD & Klug A (1962) Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 27:1–24. Doolittle RF (1995) The multiplicity of domains in proteins. Annu Rev Biochem 64:287–314. Eisenberg D (2003) The discovery of the alpha-­‐helix and beta-­‐sheet, the principle structural features of proteins. Proc Natl Acad Sci USA 100:11207–11210. Fraenkel-­‐Conrat H & Williams RC (1955) Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc Natl Acad Sci USA 41:690–698. Goodsell DS & Olson AJ (2000) Structural symmetry and protein function. Annu Rev Biophys Biomol Struct 29:105–153. Harrison SC (1992) Viruses. Curr Opin Struct Biol 2:293– 299. Harrison SC (2004) Whither structural biology? Nature Struct Mol Biol 11:12–15. Hudder A, Nathanson L & Deutscher MP (2003) Organization of mammalian cytoplasm. Mol Cell Biol 23:9318–9326. International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. Meiler J & Baker D (2003) Coupled prediction of protein secondary and tertiary structure. Proc Natl Acad Sci USA 100:12105–12110. Nomura M (1973) Assembly of bacterial ribosomes. Science 179: 864–873. Orengo CA & Thornton JM (2005) Protein families and their evolution— a structural perspective. Annu Rev Biochem 74:867–900. Pauling L & Corey RB (1951) Configurations of polypeptide chains with favored orientations around single bonds: two new pleated sheets. Proc Natl Acad Sci USA 37:729–740. Pauling L, Corey RB & Branson HR (1951) The structure of proteins: two hydrogen-­‐bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci USA 37:205–211. Essential Cell Biology, Fourth Edition REFERENCES Ponting CP, Schultz J, Copley RR et al. (2000) Evolution of domain families. Adv Protein Chem 54:185–244. Reynolds KA, Mc Laughlin RN, Ranganathan R (2011) Hot spots for allosteric regulation on protein surfaces. Cell 147(7): 1564-­‐75. Trinick J (1992) Understanding the functions of titin and nebulin. FEBS Lett 307:44–48. Vogel C, Bashton M, Kerrison ND et al (2004) Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol 14:208–216. Zhang C & Kim SH (2003) Overview of structural genomics: from structure to function. Curr Opin Chem Biol 7:28–32. Protein Function Alberts B (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92:291–294. Benkovic SJ (1992) Catalytic antibodies. Annu Rev Biochem 61:29–54. Berg OG & von Hippel PH (1985) Diffusion-­‐controlled macromolecular interactions. Annu Rev Biophys Biophys Chem 14:131–160. Bhattacharyya RP, Remenyi A, Yeh BJ & Lim WA (2006) Domains, motifs, and scaffolds: The role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem 75:655–680. Bourne HR (1995) GTPases: a family of molecular switches and clocks. Philos Trans R Soc Lond B 349:283–289. Braden BC & Poljak RJ (1995) Structural features of the reactions between antibodies and protein antigens. FASEB J 9:9–16. Dickerson RE & Geis I (1983) Hemoglobin: Structure, Function, Evolution and Pathology. Menlo Park, CA: Benjamin Cummings. Dressler D & Potter H (1991) Discovering Enzymes. New York: Scientific American Library. Eisenberg D, Marcotte, EM, Xenarios, I & Yeates TO (2000) Protein function in the post-­‐genomic era. Nature 405:823–826. Fersht AR (1999) Structure and Mechanisms in Protein Science: A Guide to Enzyme Catalysis. New York: WH Freeman. Johnson, LN & Lewis RJ (2001) Structural basis for control by phosphorylation. Chem Rev 101:2209– 2242. Kantrowitz ER & Lipscomb WN (1988) Escherichia coli aspartate transcarbamoylase: the relation between structure and function. Science 241:669–674. Khosla C & Harbury PB (2001) Modular enzymes. Nature 409:247–252. Kim E & Sheng M (2004) PDZ domain proteins of synapses. Nature Rev Neurosci 5:771–781. Koshland DE, Jr (1984) Control of enzyme activity and metabolic pathways. Trends Biochem Sci 9:155–159. Kraut DA, Carroll KS & Herschlag D (2003) Challenges in enzyme mechanism and energetics. Annu Rev Biochem 72:517–571. Krogan NJ, Cagney G, Yu H et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisisae. Nature 440:637–643. Lichtarge O, Bourne HR & Cohen FE (1996) An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257:342–358. Marcotte EM, Pellegrini M, Ng HL et al (1999) Detecting protein function and protein–protein interactions from genome sequences. Science 285:751–753. Monod J, Changeux JP & Jacob F (1963) Allosteric proteins and cellular control systems. J Mol Biol 6:306–329. Pawson T & Nash P (2003) Assembly of regulatory systems through protein interaction domains. Science 300:445–452. Pavletich NP (1999) Mechanisms of cyclin-­‐dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 287:821–828. Pellicena P & Kuriyan J (2006) Protein-­‐protein interactions in the allosteric regulation of protein kinases. Curr Opin Struct Biol 16:702–709. Perutz M (1990) Mechanisms of Cooperativity and Allosteric Regulation in Proteins. Cambridge: Cambridge University Press. Raushel F, Thoden, JB & Holden HM (2003) Enzymes with molecular tunnels. Acc Chem Res 36:539–548. Radzicka A & Wolfenden R (1995) A proficient enzyme. Science 267:90–93. Sato TK, Overduin M & Emr S (2001) Location, location, location: Membrane targeting directed by PX domains. Science 294:1881–1885. Schramm VL (1998) Enzymatic transition states and transition state analog design. Annu Rev Biochem 67:693–720. Schultz PG & Lerner RA (1995) From molecular diversity to catalysis: lessons from the immune system. Science 269:1835–1842. Vale RD & Milligan RA (2000) The way things move: looking under the hood of molecular motor proteins. Science 288:88–95. Vocadlo DJ, Davies GJ, Laine R & Withers SG (2001) Catalysis by hen egg-­‐white lysozyme proceeds via a covalent intermediate. Nature 412:835–838. Essential Cell Biology, Fourth Edition REFERENCES Walsh C (2001) Enabling the chemistry of life. Nature 409:226–231. Yang XJ (2005) Multisite protein modification and intramolecular signaling. Oncogene 24:1653–1662. Zhu H, Bilgin M & Snyder M (2003) Proteomics. Annu Rev Biochem 72:783–812. Purifying Proteins de Duve C & Beaufay H (1981) A short history of tissue fractionation. J Cell Biol 91:293s–299s. Krogan NJ, Cagney G, Yu H et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–43. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. Nirenberg MW & Matthaei JH (1961) The dependence of cell-­‐free protein synthesis in E. coli on naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci. USA 47:1588–1602. O’Farrell PH (1975) High-­‐resolution two-­‐dimensional electrophoresis of proteins. J Biol Chem 250:4007– 4021. Palade G (1975) Intracellular aspects of the process of protein synthesis. Science 189:347–358. Scopes RK & Cantor CR (1994) Protein Purification: Principles and Practice, 3rd ed. New York: Springer-­‐ Verlag. Strucutral Genomics Consortium et al (2008) Protein production and purification. Nat Methods 5(2): 135-­‐ 46. Analyzing Proteins Branden C & Tooze J (1999) Introduction to Protein Structure, 2nd ed. New York: Garland Science. Fields S & Song O (1989) A novel genetic system to detect protein–protein interactions. Nature 340:245–246. Giepmans BN, Adams SR et al (2006) The fluorescent toolbox for assessing protein location and function. Science 312:217–24. Kendrew JC (1961) The three-­‐dimensional structure of a protein molecule. Sci Am 205:96–111. Knight ZA & Shokat KM (2007) Chemical genetics: Where genetics and pharmacology meet. Cell 128:425–30. Rigaut G, Shevchenko A, Rutz B et al (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol 17:1030–1032. Washburn MP, Wolters D and Yates JR (2001) Large-­‐ scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnol 19:242–7. Wuthrich K (1989) Protein structure determination in solution by nuclear magnetic resonance spectroscopy. Science 243:45–50. Chapter 5 The Structure and Function of DNA Avery OT, MacLeod CM & McCarty M (1944) Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med 79:137–158. Meselson M & Stahl FW (1958) The replication of DNA in E. coli. Proc Natl Acad Sci USA 44:671–682. Watson JD & Crick FHC (1953) Molecular structure of nucleic acids. A structure for deoxyribose nucleic acids. Nature 171:737–738. Chromosomal DNA and Its Packaging in the Chromatin Fiber Jin J, Cai Y, Li B et al (2005) In and out: histone variant exchange in chromatin. Trends Biochem Sci 30:680– 687. Kornberg RD & Lorch Y (1999) Twenty-­‐five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294. Li G, Levitus M, Bustamante C & Widom J (2005) Rapid spontaneous accessibility of nucleosomal DNA. Nature Struct Mol Biol 12:46–53. Lorch Y, Maier-­‐Davis B & Kornberg RD (2006) Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci USA 103:3090–3093. Luger K, Mader AW, Richmond RK et al (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–260. Luger K & Richmond TJ (1998) The histone tails of the nucleosome. Curr Opin Genet Dev 8:140–146. Malik H S & Henikoff S (2003) Phylogenomics of the nucleosome Nature Struct Biol 10:882-­‐891. Ried T, Schrock E, Ning Y & Wienberg J (1998) Chromosome painting: a useful art. Hum Mol Genet 7:1619–1626. Robinson PJ & Rhodes R (2006) Structure of the 30 nm chromatin fibre: A key role for the linker histone. Curr Opin Struct Biol 16:1–8. Saha A, Wittmeyer J & Cairns BR (2006) Chromatin remodeling: the industrial revolution of DNA around histones. Nature Rev Mol Cell Biol 7:437–446. Woodcock CL (2006) Chromatin architecture. Curr Opin Struct Biol 16:213–220. Essential Cell Biology, Fourth Edition REFERENCES The Regulation of Chromatin Structure Burgess RJ, Zhang Z (2010) Histones, histone chaperone and nucleosome assembly. Protein Cell 1(7): 607-­‐12. Egger G, Liang, G, Aparicio A & Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463. Henikoff S (1990) Position-­‐effect variegation after 60 years. Trends Genet 6:422–426. Henikoff S & Ahmad K (2005) Assembly of variant histones into chromatiin. Annu Rev Cell Dev Biol 21:133–153. Gaszner M & Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Rev Genet 7:703–713. Hake SB & Allis CD (2006) Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis.” Proc Natl Acad Sci USA 103:6428–6435. Jenuwein T (2006) The epigenetic magic of histone lysine methylation. FEBS J 273:3121–3135. Martin C & Zhang Y (2005) The diverse functions of histone lysine methylation. Nature Rev Mol Cell Biol 6:838–849. Mellone B, Erhardt S & Karpen GH (2006) The ABCs of centromeres. Nature Cell Biol 8:427–429. Peterson CL & Laniel MA (2004) Histones and histone modifications. Curr Biol 14:R546–R551. Ruthenburg AJ, Allis CD & Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25:15–30. Shahbazian MD & Grunstein M (2007) Functions of site-­‐specific histone acetylation and deacetylation. Annu Rev Biochem 76:75–100. Zentner GE, Henikoff S (2013) Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20(3): 259-­‐66. The Global Structure of Chromosomes Akhtar A & Gasser SM (2007) The nuclear envelope and transcriptional control. Nature Rev Genet 8:507– 517. Callan HG (1982) Lampbrush chromosomes. Proc Roy Soc Lond Ser B 21:417–448. Chakalova L, Debrand E, Mitchel JA et al (2005) Replication and transcription: shaping the landscape of the genome. Nature Rev Genet 6:669–678. Cremer T, Cremer M, Dietzel S et al (2006) Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol 18:307–316. Ebert A, Lein S, Schotta G & Reuter G (2006) Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res 14:377–392. Fraser P & Bickmore W (2007) Nuclear organization of the genome and the potential for gene regulation. Nature 447:413–417. Handwerger KE & Gall JG (2006) Subnuclear organelles: new insights into form and function. Trends Cell Biol 16:19–26. Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nature Rev Mol Cell Biol 7:311– 322. Lamond AI & Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nature Rev Mol Cell Biol 4:605–612. Maeshima K & Laemmli UK (2003) A two-­‐step scaffolding model for mitotic chromosome assembly. Dev Cell 4:467–480. Sims JK, Houston SI, Magazinnik T & Rice JC (2006) A trans-­‐tail histone code defined by monomethylated H4 Lys-­‐20 and H3 Lys-­‐9 demarcates distinct regions of silent chromatin. J Biol Chem 281:12760–12766. Speicher MR & Carter NP (2005) The new cytogenetics: blurring the boundaries with molecular biology. Nature Rev Genet 6:782–792. Zhimulev IF (1998) Morphology and structure of polytene chromosomes. Adv Genet 37:1–566. Chapter 6 The Maintenance of DNA Sequences Cooper GM, Brudno M, Stone ES et al (2004) Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res 14:539–548. Crow JF (2000) The origins, patterns and implications of human spontaneous mutation. Nature Rev Genet 1:40–47. Hedges SB (2002) The origin and evolution of model organisms. Nature Rev Genet 3:838–849. King MC, Wilson AC (1965) Evolution at two levels in humans and chimpanzees. Science 188:107–16. DNA Replication Mechanisms Alberts B (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92:291–294. Dillingham MS (2006) Replicative helicases: a staircase with a twist. Curr Biol 16:R844–R847. Indiani C & O’Donnell M (2006) The replication clamp-­‐ loading machine at work in the three domains of life. Nature Rev Mol Cell Biol 7:751–761. Essential Cell Biology, Fourth Edition REFERENCES Kornberg A (1960) Biological synthesis of DNA. Science 131:1503–1508. Li JJ & Kelly TJ (1984) SV40 DNA replication in vitro. Proc Natl Acad Sci USA 81:6973. Meselson M & Stahl FW (1958) The replication of DNA in E. coli. Proc Natl Acad Sci USA 44:671–682. Modrich P & Lahue R (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem 65:101–133. Mott ML & Berger JM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nature Rev Microbiol 5:343–354. O’Donnell M (2006) Replisome architecture and dynamics in E. coli. J Biol Chem 281:10653–10656. Okazaki R, Okazaki T, Sakabe K et al. (1968) Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci USA 59:598–605. Raghuraman MK, Winzeler EA, Collingwood D et al (2001) Replication dynamics of the yeast genome. Science 294:115–121. Rao PN & Johnson RT (1970) Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature 225:159. Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev Mol Cell Biol 3:430–440. The Initiation and Completion of DNA Replication in Chromosomes Aves SJ (2009) DNA replication initiation. Methods Mol Biol 521:3-­‐17 Chan SR & Blackburn EH (2004) Telomeres and telomerase. Philos Trans R Soc Lond B Bio Sci 359:109–121. Costa S & Blow JJ (2007) The elusive determinants of replication origins. EMBO Rep 8:332–334. Groth A, Rocha W & Almouzni G (2007) Chromatin challenges during DNA replication and repair. Cell 128:721–733. Machida YJ, Hamlin JL & Dutta A (2005) Right place, right time, and only once: replication initiation in metazoans. Cell 123:13–24. O’Donnell M & Kuriyan J (2005) Clamp loaders and replication initiation. Curr Opin Struct Biol 16:35–41. O’Sullivant RJ and KArlseder J (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11(3): 171-­‐81. Murnane JP (2012) Telomere dysfunction and chromosome instability. Mutat Res 730(1-­‐2): 28-­‐36. Robinson NP & Bell SD (2005) Origins of DNA replication in the three domains of life. FEBS J 272:3757–3766. Smogorzewska A & de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73:177–208. DNA Repair Barnes DE, & Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38:445–476. Harrison JC & Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40:209–235. Heller RC & Marians KJ (2006) Replisome assembly and the direct restart of stalled replication. Nature Rev Mol Cell Biol 7:932–43. Lieber M, Ma Y et al (2003) Mechanism and regulation of human nonhomologous DNA end-­‐joining. Nature Rev Mol Cell Biol 4:712–720. Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715. Prakash S & Prakash L (2002) Translesion DNA synthesis in eukaryotes: a one-­‐ or two-­‐polymerase affair. Genes Dev 16:1872–1883. Sancar A, Lindsey-­‐Boltz LA et al. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85. Svejstrup JQ (2002) Mechanisms of transcription-­‐ coupled DNA repair. Nature Rev Mol Cell Biol 3:21– 29. Wyman C & Kanaar R (2006) DNA double-­‐strand break repair: all’s well that ends well. Annu Rev Genet 40:363–383. Homologous Recombination Adams MD, McVey M & Sekelsky JJ (2003) Drosophila BLM in double-­‐strand break repair by synthesis-­‐ dependent strand annealing. Science 299:265–267. Cox MM (2001) Historical overview: searching for replication help in all of the rec places. Proc Natl Acad Sci USA 98:8173–8180. Holliday R (1990) The history of the DNA heteroduplex. BioEssays 12:133–142. Lisby M, Bartow JH et al (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699– 713. Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNa damage tolerance. Cell Res 18(1): 99-­‐113. McEachern MJ & Haber JE (2006) Break-­‐induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75:111–135. Essential Cell Biology, Fourth Edition REFERENCES Michel B, Gromponee G et al (2004) Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA 101:12783–12788. Szostak JW, Orr-­‐Weaver TK, Rothstein RJ et al (1983) The double-­‐strand break repair model for recombination. Cell 33:25–35. West SC (2003) Molecular views of recombination proteins and their control. Nature Rev Mol Cell Biol 4:435–445. Chapter 7 From DNA to RNA Adams RL, Wente SR (2013) Uncovering nuclear pore complexity with innovation. Cell 152(6): 1218-­‐21. Bentley DL (2005) Rules of engagement: co-­‐ transcriptional recruitment of pre-­‐mRNA processing factors. Curr Opin Cell Biol 17:251–256. Berget SM, Moore C & Sharp PA (1977) Spliced segments at the 5¢ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA 74:3171–3175. Black DL (2003). Mechanisms of alternative pre-­‐ messenger RNA splicing. Annu Rev Biochem 72:291– 336. Brenner S, Jacob F & Meselson M (1961) An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190:576– 581. Cate JH, Gooding AR, Podell E et al. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273:1678–1685. Chow LT, Gelinas RE, Broker TR et al (1977) An amazing sequence arrangement at the 5¢ ends of adenovirus 2 messenger RNA. Cell 12:1–8. Cramer P (2002) Multisubunit RNA polymerases. Curr Opin Struct Biol 12:89–97. Daneholt B (1997) A look at messenger RNP moving through the nuclear pore. Cell 88:585–588. Dreyfuss G, Kim VN, & Kataoka N (2002) Messenger-­‐ RNA-­‐binding proteins and the messages they carry. Nature Rev Mol Cell Biol 3:195–205. Houseley J, LaCava J & Tollervey D (2006) RNA-­‐quality control by the exosome. Nature Rev Mol Cell Biol 7:529–539. Izquierdo JM, & Valcárcel J (2006) A simple principle to explain the evolution of pre-­‐mRNA splicing. Genes Dev 20:1679–1684. Kornberg RD (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30:235–239. Malik S & Roeder RG (2005) Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30:256–263. Matsui T, Segall J, Weil PA & Roeder RG (1980) Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem 255:11992–11996. Patel AA & Steitz JA (2003) Splicing double: insights from the second spliceosome. Nature Rev Mol Cell Biol 4|:960–970. Phatnani HP & Greenleaf AL (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20:2922–2936. Query CC & Konarska MM (2006) Splicing fidelity revisited. Nature Struct Mol Biol 13:472–474. Ruskin B, Krainer AR, Maniatis T et al (1984) Excision of an intact intron as a novel lariat structure during pre-­‐ mRNA splicing in vitro. Cell 38:317–331. Spector DL (2003) The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72:573–608. Staley JP & Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315–326. Thomas MC & Chiang CM (2006) The general transcription machinery and general cofactors. Critical Rev Biochem Mol Biol 41:105–178. Wang D, Bushnell DA, Westover KD et al (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127:941–954. From RNA to Protein Allen GS & Frank J (2007) Structural insights on the translation initiation complex: ghosts of a universal initiation complex. Mol Microbiol 63:941–950. Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230. Brunelle JL, Youngman EM, Sharma D et al (2006) The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA 12:33–39. Chien P, Weissman JS, & DePace AH (2004). Emerging principles of conformation-­‐based prion inheritance. Annu Rev Biochem 73:617–656. Crick FHC (1966) The genetic code: III. Sci Am 215:55– 62. Da Fonseca PC, He J, Morriss EP (2012) Molecular model of the human 26S proteasome. Mol Cell 46:54-­‐66. Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79:351-­‐79. Essential Cell Biology, Fourth Edition REFERENCES Hershko A, Ciechanover A & Varshavsky A (2000) The ubiquitin system. Nature Med 6:1073–1081. Ibba M & Soll D (2000) Aminoacyl-­‐tRNA synthesis. Annu Rev Biochem 69:617–650. Kornblihtt AR et al. (2013) Alternative splicing: a pivitorl step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14(3): 153-­‐165. Kozak M (1992) Regulation of translation in eukaryotic systems. Annu Rev Cell Biol 8:197–225. Kuzmiak HA, & Maquat LE (2006) Applying nonsense-­‐ mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med 12:306– 316. Moore PB & Steitz TA (2005) The ribosome revealed. Trends Biochem Sci 30:281–283. Noller HF (2005) RNA structure: reading the ribosome. Science 309:1508–1514. Ogle JM, Carter AP & Ramakrishnan V (2003) Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci 28:259–266. Prusiner SB (1998) Nobel lecture. Prions. Proc Natl Acad Sci USA 95:13363–13383. Rehwinkel J, Raes J & Izaurralde E (2006) Nonsense-­‐ mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci 31:639–646. Sauer RT, Bolon DN, Burton BM et al (2004) Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119:9–18. Shorter J & Lindquist S (2005) Prions as adaptive conduits of memory and inheritance. Nature Rev Genet 6:435–450. Varshavsky A (2005) Regulated protein degradation. Trends in Biochem Sci 30:283–286. Voges D, Zwickl P & Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68:1015– 1068. Weissmann C (2005) Birth of a prion: spontaneous generation revisited. Cell 122:165–168. Young JC, Agashe VR, Siegers K et al (2004) Pathways of chaperone-­‐mediated protein folding in the cytosol. Nature Rev Mol Cell Biol 5:781–791. The RNA World and the Origins of Life Joyce GF (1992) Directed molecular evolution. Sci Am 267:90–97. Orgel L (2000) Origin of life. A simpler nucleic acid. Science 290:1306–1307. Kruger K, Grabowski P, Zaug P et al (1982) Self-­‐splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening seuence of Tetrahymena. Cell 31:147–157. Silverman SK (2003) Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA 9:377–383. Szostak JW, Bartel DP & Luisi PL (2001) Synthesizing life. Nature 409:387–390. Chapter 8 An Overview of Gene Control Campbell KH, McWhir J, Ritchie WA & Wilmut I (1996) Sheep cloned by nuclear transfer from a cultured cell line. Nature 380:64–66. Davidson EH (2006) The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Burlington, MA: Elsevier. Gurdon JB (1992) The generation of diversity and pattern in animal development. Cell 68:185–199. Levine M & Tjian R (2003) Transcription regulation and animal diversity. Nature 424:147–151. Roberts TC, Morris KV, Weinberg MS (2013) Perspectives on the mechanism of transcriptional regulation by long non-­‐coding RNAs. Epigenetics (9)1. Ross DT, Scherf U, Eisen MB et al (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet 24:227–235. Marro S et al. (2011) Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9:374-­‐378. DNA-­‐binding Motifs in Gene Regulatory Proteins Gehring WJ, Affolter M & Burglin T (1994) Homeodomain proteins. Annu Rev Biochem 63:487– 526. Harbison CT, Gordon DB, Lee TI et al (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104. Luscombe NM, Austin SE, Berman HM, et al (2000) An overview of the structures of protein–DNA complexes. Gen Biol 1:reviews 001.1–001.37. McKnight SL (1991) Molecular zippers in gene regulation. Sci Am 264:54–64. Pabo CO & Sauer RT (1992) Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61:1053–1095. Rhodes D & Klug A (1993) Zinc fingers. Sci Am 268:56– 65. Seeman NC, Rosenberg JM & Rich A (1976) Sequence-­‐ specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci USA 73:804–808. Essential Cell Biology, Fourth Edition REFERENCES How Genetic Switches Work Becker PB & Hörz W (2002) ATP-­‐dependent nucleosome remodeling. Annu Rev Biochem 71:247– 273. Beckwith J (1987) The operon: an historical account. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology (Neidhart FC, Ingraham JL, Low KB et al eds), vol 2, pp 1439–1443. Washington, DC: ASM Press. Gaszner M & Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Rev Genet 7:703–713. Gilbert W and Müller-­‐Hill B (1967) The lac operator is DNA. Proc Natl Acad Sci USA 58:2415. Green MR (2005) Eukaryotic transcription activation: right on target. Mol Cell 18:399–402. Jacob F & Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3:318–356. Lawson CL, Swigon D, Murakami KS et al (2004) Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol 14:10– 20. Millar CB, & Grunstein M (2006) Genome-­‐wide patterns of histone modifications in yeast. Nature Rev Mol Cell Biol 7:657–666. Narlikar GJ, Fan HY & Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487. Oehler S, Eismann ER, Krämer H et al (1990) The three operators of the lac operon cooperate in repression. EMBO J 9:973–979. Ptashne M (2004) A Genetic Switch: Phage and Lambda Revisited, 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. Ptashne M (1967) Specific binding of the lambda phage repressor to lambda DNA. Nature 214:232–234. St Johnston D & Nusslein-­‐Volhard C (1992) The origin of pattern and polarity in the Drosophila embryo. Cell 68:201–219. Strahl BD & Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45. The Molecular Genetic Mechanisms that Create Specialized Cell Types Alon, U (2006) An Introduction to Systems Biology: Design Principles of Biological Circuits (Chapman & Hall/Crc Mathematical and Computational Biology Series) TF Chapman. Bell-­‐Pedersen D, Cassone VM, Earnest DJ et al (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nature Rev Genet 6:544– 556. Bernstein BE, Meissner A & Lander ES (2007) The mammalian epigenome. Cell 128:669–681. Herskowitz I (1989) A regulatory hierarchy for cell specialization in yeast. Nature 342:749–757. Klose RJ & Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31:89–97. Meyer BJ (2000) Sex in the worm: counting and compensating X-­‐chromosome dose. Trends Genet 16:247–253. Surani MA (2001) Reprogramming of genome function through epigenetic inheritance. Nature 414:122– 128. Tapscott SJ (2005) The circuitry of a master switch: MyoD and the regulation of skeletal muscle gene transcription. Development 132:2685–2695. Post-­‐transcriptional Controls Bass BL (2002) RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71:817–846. Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126:37–47. Brennecke J, Stark A, Russell RB et al (2005) Principles of microRNAtarget recognition. PLoS Biology 3. Fire A, Xu S, Montgomery MK et al (1998) Potent and specific genetic interference by double-­‐stranded RNA in Caenorhabditis elegans. Nature 391:806–811. Frankel AD & Young JAT (1998) HIV-­‐1: fifteen proteins and an RNA. Annu Rev Biochem 67:1–25. Gottesman S (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 58:303–328. Khavari DA, Sen GL, Rinn JK (2010) DNA methylation and epigenetic control of cellular differentiation Cell Cycle 9(19): 3880-­‐3. Mello CC & Conte D (2004) Revealing the world of RNA interference. Nature 431:338–342. Parker R & Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25:635– 646. Serganov A, Nudler E (2013) A decade of riboswitches. Cell 152(1-­‐2):17-­‐24. Stuart KD, Schnaufer A, Ernst NL et al (2005) Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30:97–105. Tolia NH & Joshua-­‐Tor L (2007) Slicer and the argonautes. Nature Chem Biol 3:36–43. Tomari Y & Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19:517–529. Valencia-­‐Sanchez MA, Liu J, Hannon GJ et al (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20:515–524. Essential Cell Biology, Fourth Edition REFERENCES Verdel A & Moazed D (2005) RNAi-­‐directed assembly of heterochromatin in fission yeast. FEBS Letters 579:5872–5878. Wilhelm JE & Smibert, CA (2005) Mechanisms of translational regulation in Drosophila. Biol Cell 97:235–252. Winkler WC & Breaker RR (2005) Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59:487–517. Yates LA, Norbury CJ, Gilbert RJ (2013) The long and short of microRNA. Cell 153(3): 516-­‐9. Chapter 9 How Genomes Evolve Alfodi J, Lindblad-­‐Toh K, (2013) Comparative genomics as a tool to understand evolution and disease. Genome Res 23(7): 1063-­‐8. Batzer MA & Deininger PL (2002) ALU repeats and human genomic diversity. Nature Rev Genet 3:370– 379. Blanchette M, Green ED, Miller W, & Haussler D (2004) Reconstructing large regions of an ancestral mammalian genome in silico. Genome Res 14:2412– 2423. Cheng Z, Ventura M, She X et al (2005) A genome-­‐wide comparison of recent chimpanzee and human segmental duplications. Nature 437:88–93. Feuk L, Carson AR & Scherer S (2006) Structural variation in the human genome. Nature Rev Genet 7:85–97. International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. International Human Genome Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431:931–945. Koszul R, Caburet S, Dujon B & Fischer G (2004) Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J 23:234–243. Margulies EH, NISC Comparative Sequencing Program & Green ED (2003) Detecting highly conserved regions of the human genome by multispecies sequence comparisons. Cold Spring Harbor Symp Quant Biol 68:255–263. Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. Pollard KS, Salama SR, Lambert N et al (2006) An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443:167–172. Sharp AJ, Cheng Z & Eichler EE (2007) Structural variation of the human genome. Annu Rev Genomics Hum Genet 7:407–442. Siepel A, Bejerano G, Pedersen JS et al (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15:1034–1050. The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320. The ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816. The Diversity of Genomes and the Tree of Life Blattner FR, Plunkett G, Bloch CA et al (1997) The complete genome sequence of Escherichia coli K-­‐12. Science 277:1453–1474. Boucher Y, Douady CJ, Papke RT et al (2003) Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet 37:283–328. Cole ST, Brosch R, Parkhill J et al (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. Dixon B (1994) Power Unseen: How Microbes Rule the World. Oxford: Freeman. Kerr RA (1997) Life goes to extremes in the deep earth—and elsewhere? Science 276:703–704. Lee TI, Rinaldi NJ, Robert F et al (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799–804. Olsen GJ & Woese CR (1997) Archaeal genomics: an overview. Cell 89:991–994. Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740. Woese C (1998) The universal ancestor. Proc Natl Acad Sci USA 95:6854–6859. Genetic Information in Eukaryotes Adams MD, Celniker SE, Holt RA et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195. Andersson SG, Zomorodipour A, Andersson JO et al (1998) The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133–140. The Arabidopsis Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. Carroll SB, Grenier JK & Weatherbee SD (2005) From DNA to Diversity: Molecular Genetics and the Essential Cell Biology, Fourth Edition REFERENCES Evolution of Animal Design, 2nd ed. Maldon, MA: Blackwell Science. de Duve C (2007) The origin of eukaryotes: a reappraisal. Nature Rev Genet 8:395-­‐403. Delsuc F, Brinkmann H & Philippe H (2005) Phylogenomics and the reconstruction of the tree of life. Nature Rev Genet 6:361–375. DeRisi JL, Iyer VR & Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. Gabriel SB, Schaffner SF, Nguyen H et al (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229. Goffeau A, Barrell BG, Bussey H et al (1996) Life with 6000 genes. Science 274:546–567. International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. Kellis M, Birren BW & Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617–624. Lynch M & Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151– 1155. Mulley J & Holland P (2004) Comparative genomics: Small genome, big insights. Nature 431:916–917. National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/ Owens K & King MC (1999) Genomic views of human history. Science 286:451–453. Palmer JD & Delwiche CF (1996) Second-­‐hand chloroplasts and the case of the disappearing nucleus. Proc Natl Acad Sci USA 93:7432–7435. Pennisi E (2004) The birth of the nucleus. Science 305:766–768. Plasterk RH (1999) The year of the worm. BioEssays 21:105–109. Reed FA & Tishkoff SA (2006) African human diversity, origins and migrations. Curr Opin Genet Dev 16:597– 605. Rubin GM, Yandell MD, Wortman JR et al (2000) Comparative genomics of the eukaryotes. Science 287:2204–2215. Stillman B & Stewart D (2003) The genome of Homo sapiens. (Cold Spring Harbor Symp. Quant. Biol. LXVIII). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018. Tinsley RC & Kobel HR (eds) (1996) The Biology of Xenopus. Oxford: Clarendon Press. Tyson JJ, Chen KC & Novak B (2003) Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol 15:221–231. Venter JC, Adams MD, Myers EW et al (2001) The sequence of the human genome. Science 291:1304– 1351. Transposition and Site-­‐Specific Recombination Ade C, Roy-­‐Engel AM, Deininger PL (2013) Alu elements: an intrinsic source of human genome instability. Curr Opin Virol pii: S1879-­‐6257(13)00153-­‐ 3. Botstein D, White RL, Skolnick M & Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331. Campbell AM (1993) Thirty years ago in genetics: prophage insertion into bacterial chromosomes. Genetics 133:433–438. Comfort NC (2001) From controlling elements to transposons: Barbara McClintock and the Nobel Prize. Trends Biochem Sci 26:454–457. Craig NL (1996) Transposition, in Escherichia coli and Salmonella, pp 2339–2362. Washington, DC: ASM Press. Frost, LS (2005) Mobile genetic elemnts: the agents of open source evolution. Nat Rev Microbiol 3(9): 722-­‐ 32. Gottesman M (1999) Bacteriophage lambda: the untold story. J Mol Biol 293:177–180. Grindley ND, Whiteson KL & Rice PA (2006) Mechanisms of site-­‐specific recombination. Annu Rev Biochem 75:567–605. International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–320. Malachowa N, DeLeo FR (2010) Mobile genetic elements of Staphylococcus aureas. Cell Mol Life Sci 67(18): 3057-­‐71 Rebollo R, Romanish MT, Mager DL (2012) Transposable elemnts: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21-­‐26. Rubin GM & Sprading AC (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218:348–353. Sabeti PC, Schaffner SF, Fry B et al (2006) Positive natural selection in the human lineage. Science 312:1614–1620. Varmus H (1988) Retroviruses. Science 240:1427–1435. Zickler D & Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33:603–754. Essential Cell Biology, Fourth Edition REFERENCES Chapter 10 Isolating Cells and Growing Them in Culture Emmert-­‐Buck MR, Bonner RF, Smith PD et al (1996) Laser capture microdissection. Science 274:998– 1001. Ham RG (1965) Clonal growth of mammalian cells in a chemically defined, synthetic medium. Proc Natl Acad Sci USA 53:288–293. Harlow E & Lane D (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. Herzenberg LA, Sweet RG & Herzenberg LA (1976) Fluorescence-­‐activated cell sorting. Sci Am 234:108– 116. Levi-­‐Montalcini R (1987) The nerve growth factor thirty-­‐five years later. Science 237:1154–1162. Lerou PH & Daley GQ (2005) Therapeutic potential of embryonic stem cells. Blood Rev 19:321–31. Milstein C (1980) Monoclonal antibodies. Sci Am 243:66–74. Isolating, Cloning, and Sequencing DNA Adams MD, Celniker SE, Holt RA et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195. Alwine JC, Kemp DJ & Stark GR (1977) Method for detection of specific RNAs in agarose gels by transfer to diabenzyloxymethyl paper and hybridization with DNA probes. Proc Natl Acad Sci USA 74:5350–5354. Blattner FR, Plunkett G, Bloch CA et al (1997) The complete genome sequence of Escherichia coli K-­‐12. Science 277:1453–1474. Cohen S, Chang A, Boyer H & Helling R (1973) Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci USA 70:3240– 3244. Hert DG, Fredlake CP, Barron AE (2008) Advantages and limitations of next-­‐generation sequencing technologies: a comparison of electrophoresis and non-­‐electrophoresis methods. Electrophoresis 29(23):4618-­‐26. International Human Genome Sequencing Consortium (2000) Initial sequencing and analysis of the human genome. Nature 409:860–921. International Human Genome Sequencing Consortium (2006) The DNA sequence, annotation and analysis of human chromosome 3. Nature 440:1194–1198. Jackson D, Symons R & Berg P (1972) Biochemical method for inserting new genetic information into DNA of simian virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Natl Acad Sci USA 69:2904–2909. Maniatis T et al (1978) The isolation of structural genes from libraries of eukaryotic DNA. Cell 15:687–701. Mardis ER (2013) Next-­‐generation sequencing platofrms. Annu Rev Anal Chem (Palo Alto Calif) 6:287-­‐303. Mullis KB (1990). The unusual origin of the polymerase chain reaction. Sci Am 262:56–61. Nathans D & Smith HO (1975) Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem 44:273–93. Saiki RK, Gelfand DH, Stoffel S et al (1988) Primer-­‐ directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487– 491. Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. Sanger F, Nicklen S & Coulson AR (1977) DNA sequencing with chain-­‐terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467. Smith M (1994) Nobel lecture. Synthetic DNA and biology. Biosci Rep 14:51–66. Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517. The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018. Venter JC, Adams MA, Myers EW et al (2000) The sequence of the human genome. Science 291:1304– 1351. Studying Gene Expression and Function Boone C, Bussey H & Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nature Rev Genet 8:437–449. DeRisi JL, Iyer VR & Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. Hawrylycz M et al. (2011) Digital atlasing and standardization in the mouse brain. PLoS Comput Biol 7:e1001065. Jinek M, Doudna JA (2009) A three-­‐dimensional view of the molecular machinery of RNA interference. Nature (7228): 405-­‐12. Essential Cell Biology, Fourth Edition REFERENCES Livet J et al. (2007) Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56-­‐62. Lockhart DJ & Winzeler EA (2000) Genomics, gene expression and DNA arrays. Nature 405:827–836. Mello CC & Conte D (2004) Revealing the world of RNA interference. Nature 431:338–342. Nusslein-­‐Volhard C & Weischaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801. Palmiter RD & Brinster RL (1985) Transgenic mice. Cell 41:343–345. Weigel D & Glazebrook J (2001) Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. Wilson RC, Doudna JA (2013) Molecular mechanisms of RNA interference. Annu Rev Biophys 42: 217-­‐39. Chapter 11 The Lipid Bilayer Bevers EM, Comfurius P & Zwaal RF (1999) Lipid translocation across the plasma membrane of mammalian cells. Biochim Biophys Acta 1439:317– 330. Devaux PF (1993) Lipid transmembrane asymmetry and flip-­‐flop in biological membranes and in lipid bilayers. Curr Opin Struct Biol 3:489–494. Dowhan W (1997) Molecular basis for membrane phospholipid diversity:why are there so many lipids? Annu Rev Biochem 66:199–232. Hakomori Si SI (2002) Inaugural Article:The glycosynapse. Proc Natl Acad Sci USA 99:225–232. Harder T & Simons K (1997) Caveolae, DIGs, and the dynamics of sphingolipid-­‐cholesterol microdomains. Curr Opin Cell Biol 9:534–542. Hazel JR (1995) Thermal adaptation in biological membranes:is homeoviscous adaptation the explanation? Annu Rev Physiol 57:19–42. Ichikawa S & Hirabayashi Y (1998) Glucosylceramide synthase and glycosphingolipid synthesis. Trends Cell Biol 8:198–202. Kornberg RD & McConnell HM (1971) Lateral diffusion of phospholipids in a vesicle membrane. Proc Natl Acad Sci USA 68:2564–2568. Mansilla MC, Cybulski LE & de Mendoza D (2004) Control of membrane lipid fluidity by molecular thermosensors. J Bacteriol 186:6681–6688. McConnell HM & Radhakrishnan A (2003) Condensed complexes of cholesterol and phospholipids. Biochim Biophys Acta 1610:159–73. Pomorski T & Menon AK (2006) Lipid flippases and their biological functions. Cell Mol Life Sci 63:2908– 2921. Rothman JE & Lenard J (1977) Membrane asymmetry. Science 195:743–53. Scott HL (2002) Modeling the lipid component of membranes. Curr Opin Struct Biol 12:499. Simons K & Vaz WL (2004) Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 33:269–95. Sonnino S, Prinetti A (2013) Membraine domains and the “lipid raft” concept. Curr Med Chem 20(1):4-­‐21. Tanford C (1980) The Hydrophobic Effect: Formation of Micelles and Biological Membranes. New York: Wiley. van Meer G (2005) Cellular lipidomics. EMBO J 24:3159–3165. Membrane Proteins Bennett V & Baines AJ (2001) Spectrin and ankyrin-­‐ based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev 81:1353– 1392. Bijlmakers MJ & Marsh M (2003) The on-­‐off story of protein palmitoylation. Trends Cell Biol 13:32–42. Branden C & Tooze J (1999) Introduction to Protein Structure, 2nd ed. New York: Garland Science. Bretscher MS & Raff MC (1975) Mammalian plasma membranes. Nature 258:43–49. Buchanan SK (1999) Beta-­‐barrel proteins from bacterial outer membranes:structure, function and refolding. Curr Opin Struct Biol 9:455–461. Chen Y, Lagerholm BC & Jacobson K (2006) Methods to measure the lateral diffusion of membrane lipids and proteins. Methods 39:147–153. Curranb AR & Engelman DM (2003) Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol 13:412 Deisenhofer J & Michel H (1991) Structures of bacterial photosynthetic reaction centers. Annu Rev Cell Biol 7:1–23. Drickamer K & Taylor ME (1993) Biology of animal lectins. Annu Rev Cell Biol 9:237–64. Drickamer K & Taylor ME (1998) Evolving views of protein glycosylation. Trends Biochem Sci 23:321– 324. Fiedler S, Broecker J, Keller S (2010) Protein folding in membranes. Cell Mol Life Sci 67(11): 1179-­‐98. Frye LD & Edidin M (1970) The rapid intermixing of cell surface antigens after formation of mouse-­‐human heterokaryons. J Cell Sci 7:319–335. Essential Cell Biology, Fourth Edition REFERENCES Helenius A and Simons K (1975) Solubilization of membranes by detergents. Biochim Biophys Acta 415:29–79. Henderson R & Unwin PN (1975) Three-­‐dimensional model of purple membrane obtained by electron microscopy. Nature 257:28–32. Kyte J & Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. le Maire M, Champeil P et al (2000) Interaction of membrane proteins and lipids with solubilizing detergents. Biochim Biophys Acta 1508:86–111. Lee AG (2003) Lipid-­‐protein interactions in biological membranes:a structural perspective. Biochim Biophys Acta 1612:1–40. Marchesi VT, Furthmayr H et al (1976) The red cell membrane. Annu Rev Biochem 45:667–698. Nakada C, Ritchie K & Kusumi A (2003) Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat Cell Biol 5:626–632. Oesterhelt D (1998) The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr Opin Struct Biol 8:489–500. Rath A, Deber CM (2012) Protein structure in membrane domains. Annu Rev Biophys 41:135-­‐55 Reig N & van der Goot FG (2006) About lipids and toxins. FEBS Lett 580:5572–5579. Reithmeier RAF (1993) The erythrocyte anion transporter (band 3) Curr Opin Cell Biol 7:707–714. Rodgers W & Glaser M (1993) Distributions of proteins and lipids in the erythrocyte membrane. Biochemistry 32:12591–12598. Sharon N & Lis H (2004) History of lectins:from hemagglutinins to biological recognition molecules. Glycobiology 14:53R–62R. Sheetz MP (2001) Cell control by membrane-­‐ cytoskeleton adhesion. Nat Rev Mol Cell Biol 2:392– 396. Silvius JR (1992) Solubilization and functional reconstitution of biomembrane components. Annu Rev Biophys Biomol Struct 21:323–348. Steck TL (1974) The organization of proteins in the human red blood cell membrane. A review. J Cell Biol 62:1–19. Subramaniam S (1999) The structure of bacteriorhodopsin:an emerging consensus. Curr Opin Struct Biol 9:462–468. Viel A & Branton D (1996) Spectrin:on the path from structure to function. Curr Opin Cell Biol 8:49–55. Wallin E & von Heijne G (1998) Genome-­‐wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 7:1029–1038. White SH & Wimley WC (1999) Membrane protein folding and stability:physical principles. Annu Rev Biophys Biomol Struct 28:319–365. Chapter 12 Principles of Membrane Transport Al-­‐Awqati Q (1999) One hundred years of membrane permeability: does Overton still rule? Nature Cell Biol 1: E201–E202. Cajal SR, Histologie du System Nerveux de l’Homme et de Vertebres, 1909-­‐1911. Paris: Maloine; reprinted, Madrid: CSIC, 1972. De Groot BL & Grubmuller H (2001) Water permeation across biological membranes: mechanism and dynamics of aquaporin-­‐1 and GlpF. Nature 294:2353-­‐ 7. Forrest LR & Sansom MS (2000) Membrane simulations: bigger and better? Curr Opin Struct Biol 10:174–181. Gouaux E and MacKinnon R (2005) Principles of selective ion transport in channels and pumps. Science 310:1461–1465. Mitchell P (1977) Vectorial chemiosmotic processes. Annu Rev Biochem 46:996–1005. Tanford C (1983) Mechanism of free energy coupling in active transport. Annu Rev Biochem 52:379–409. Carrier Proteins and Active Membrane Transport Almers W & Stirling C (1984) Distribution of transport proteins over animal cell membranes. J Membr Biol 77:169–186. Baldwin SA & Henderson PJ (1989) Homologies between sugar transporters from eukaryotes and prokaryotes. Annu Rev Physiol 51:459–471. Borst P & Elferink RO (2002) Mammalian ABC transporters in health and disease. Annu Rev Biochem 71:537–592. Carafoli E & Brini M (2000) Calcium pumps: structural basis for and mechanism of calcium transmembrane transport. Curr Opin Chem Biol 4:152–161. Dean M, Rzhetsky A et al (2001) The human ATP-­‐ binding cassette (ABC) transporter superfamily. Genome Res 11:1156–1166. Doige CA & Ames GF (1993) ATP-­‐dependent transport systems in bacteria and humans: relevance to cystic fibrosis and multidrug resistance. Annu Rev Microbiol 47:291–319. Essential Cell Biology, Fourth Edition REFERENCES Gadsby DC, Vergani P & Csanady L (2006) The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440:477–83. Higgins CF (2007) Multiple molecular mechanisms for multidrug resistance transporters. Nature 446:749– 57. Kaback HR, Sahin-­‐Toth M et al (2001) The kamikaze approach to membrane transport. Nature Rev Mol Cell Biol 2:610–620. Kühlbrandt W (2004) Biology, structure and mechanism of P-­‐type ATPases. Nature Rev Mol Cell Biol 5:282–295. Lodish HF (1986) Anion-­‐exchange and glucose transport proteins: structure, function, and distribution. Harvey Lect 82:19–46. Pedersen PL & Carafoli E (1987) Ion motive ATPases. 1. Ubiquity, properties, and significance to cell function. Trends Biochem Sci 12:146–150. + Romero MF & Boron WF (1999) Electrogenic Na /HCO3 cotransporters: cloning and physiology. Annu Rev Physiol 61:699–723. Saier MH, Jr (2000) Vectorial metabolism and the evolution of transport systems. J Bacteriol 182:5029– 5035. Scarborough GA (2003) Rethinking the P-­‐type ATPase problem. Trends Biochem Sci 28:581–584. Stein WD (2002) Cell volume homeostasis: ionic and nonionic mechanisms. The sodium pump in the emergence of animal cells. Int Rev Cytol 215:231– 258. Ion Channels, Nerve Signaling, and the Electrical Properties of Membranes Armstrong C (1998) The vision of the pore. Science 280:56–57. Choe S (2002) Potassium channel structures. Nature Rev Neurosci 3:115–21. Choe S, Kreusch A & Pfaffinger PJ (1999) Towards the three-­‐dimensional structure of voltage-­‐gated potassium channels. Trends Biochem Sci 24:345–349. Dugue GP, Akemann W, Knopfel T (2012) A comprehensive concept of optogenetics. Prog Brain Res 196:1-­‐28. Franks NP & Lieb WR (1994) Molecular and cellular mechanisms of general anaesthesia. Nature 367:607–614. Greengard P (2001) The neurobiology of slow synaptic transmission. Science 294:1024–30. Hille B (2001) Ionic Channels of Excitable Membranes, 3rd ed. Sunderland, MA: Sinauer. Hucho F, Tsetlin VI & Machold J (1996) The emerging three-­‐dimensional structure of a receptor. The nicotinic acetylcholine receptor. Eur J Biochem 239:539–557. Hodgkin AL & Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117:500–544. Hodgkin AL & Huxley AF (1952) Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol 116:449–472. Husson SJ, Gottschalk A, Leifer AM (2013) Optogenetic manipulation of neuroal acticity in C. elegans: from synapse to circuits and behavior. Biol Cell 105(6): 235-­‐50 Jessell TM & Kandel ER (1993) Synaptic transmission: a bidirectional and self-­‐modifiable form of cell–cell communication. Cell 72[Suppl]:1–30. Kandel ER, Schwartz JH & Jessell TM (2000) Principles of Neural Science, 4th ed. New York: McGraw-­‐Hill. Karlin A (2002) Emerging structure of the nicotinic acetylcholine receptors. Nature Rev Neurosci 3:102– 114. Katz B (1966) Nerve, Muscle and Synapse. New York: McGraw-­‐Hill. King LS, Kozono D & Agre P (2004) From structure to disease: the evolving tale of aquaporin biology. Nature Rev Mol Cell Biol 5:687–698. La Lumiere RT (2011) A new techniwue for controlling the brain: optogenetics and its potential for use in research and the clinic. Brain Stimul 4(1): 1-­‐6. Lin D et al. (2011) Functional identification of an aggression locus in the mouse hypothalamus. Nature 470:221-­‐226. MacKinnon R (2003) Potassium channels. FEBS Lett 555:62–65. Malenka RC & Nicoll RA (1999) Long-­‐term potentiation—a decade of progress? Science 285:1870–1874. Moss SJ & Smart TG (2001) Constructing inhibitory synapses. Nature Rev Neurosci 2:240–250. Neher E and Sakmann B (1992) The patch clamp technique. Sci Am 266:44–51. Nicholls JG, Fuchs PA, Martin AR & Wallace BG (2000) From Neuron to Brain, 4th ed. Sunderland, MA: Sinauer. Numa S (1987) A molecular view of neurotransmitter receptors and ionic channels. Harvey Lect 83:121– 165. Scannevin RH & Huganir RL (2000) Postsynaptic organization and regulation of excitatory synapses. Nature Rev Neurosci 1:133–141. Seeburg PH (1993) The molecular biology of mammalian glutamate receptor channels. Trends Neurosci 16:359–365. Essential Cell Biology, Fourth Edition REFERENCES Snyder SH (1996) Drugs and the Brain. New York: WH Freeman/ Scientific American Books. Stevens CF (2004) Presynaptic function. Curr Opin Neurobiol 14:341–345. Tsien RW, Lipscombe D, Madison DV et al (1988) Multiple types of neuronal calcium channels and their selective modulation. Trends Neurosci 11:431– 438. Unwin N (2003) Structure and action of the nicotinic acetylcholine receptor explored by electron microscopy. FEBS Lett 555:91–95. Chapter 13 How Cells Obtain Energy from Food Akram M (2013) Mini-­‐review on glycolysis and cancer. J Cancer Educ 28(3): 454-­‐7. Cramer WA & Knaff DB (1990) Energy Transduction in Biological Membranes. New York: Springer-­‐Verlag. Dismukes GC, Klimov VV, Baranov SV et al (2001) The origin of atmospheric oxygen on Earth: The innovation of oxygenic photosyntheis. Proc Nat Acad Sci USA 98:2170–2175. Fell D (1997) Understanding the Control of Metabolism. London: Portland Press. Flatt JP (1995) Use and storage of carbohydrate and fat. Am J Clin Nutr 61, 952S–959S. Friedmann HC (2004) From Butybacterium to E. coli: An essay on unity in biochemistry. Perspect Biol Med 47:47–66. Fothergill-­‐Gilmore LA (1986) The evolution of the glycolytic pathway. Trends Biochem Sci 11:47–51. Heinrich R, Melendez-­‐Hevia E, Montero F et al (1999) The structural design of glycolysis: An evolutionary approach. Biochem Soc Trans 27:294–298. Huynen MA, Dandekar T & Bork P (1999) Variation and evolution of the citric-­‐acid cycle: a genomic perspective. Trends Microbiol 7:281–291. Kornberg HL (2000) Krebs and his trinity of cycles. Nature Rev Mol Cell Biol 1:225–228. Krebs HA & Martin A (1981) Reminiscences and Reflections. Oxford/New York: Clarendon Press/Oxford University Press. Krebs HA (1970) The history of the tricarboxylic acid cycle. Perspect Biol Med 14:154–170. Martin BR (1987) Metabolic Regulation: A Molecular Approach. Oxford: Blackwell Scientific. McGilvery RW (1983) Biochemistry: A Functional Approach, 3rd ed. Philadelphia: Saunders. Morowitz HJ (1993) Beginnings of Cellular Life: Metabolism Recapitulates Biogenesis. New Haven: Yale University Press Newsholme EA & Stark C (1973) Regulation of Metabolism. New York: Wiley. Chapter 14 The Mitochondrion Abrahams JP, Leslie AG, Lutter R & Walker JE (1994) Structure at 2.8Å resolution of F1-­‐ATPase from bovine heart mitochondria. Nature 370:621–628. Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72:19-­‐54. Boyer PD (1997) The ATP synthase—a splendid molecular machine. Annu Rev Biochem 66:717–749. Ernster L & Schatz G (1981) Mitochondria: a historical review. J Cell Biol 91:227s–255s. Frey TG, Renken CW & Perkins GA (2002) Insight into mitochondrial structure and function from electron tomography. Biochim Biophys Acta 1555:196–203. Pebay-­‐Peyroula E & Brandolin G (2004) Nucleotide exchange in mitochondria: insight at a molecular level. Curr Opin Struct Biol 14:420–425. Madeira VM (2012) Overview of mitochondrial bioenergetics. Methods Mol Biol 810: 1-­‐6. Meier T, Polzer P, Diederichs K et al (2005) Structure of + the rotor ring of F-­‐type Na -­‐ATPase from Ilyobacter tartaricus. Science 308:659–662. Mitchell P (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-­‐osmotic type of mechanism. Nature 191:144–148. Nicholls DG (2006) The physiological regulation of uncoupling proteins. Biochim Biophys Acta 1757:459–466. Nunnari J et al. (1997) Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell 8:1233-­‐1242. Racker E & Stoeckenius W (1974) Reconstitution of purple membrane vesicles catalyzing light-­‐driven proton uptake and adenosine triphosphate formation. J Biol Chem 249:662–663. Saraste M (1999) Oxidative phosphorylation at the fin de siecle. Science 283:1488–1493. Scheffler IE (1999) Mitochondria. New York/Chichester: Wiley-­‐Liss. Stock D, Gibbons C, Arechaga I et al. (2000) The rotary mechanism of ATP synthase. Curr Opin Struct Biol 10:672–679. Weber J (2007) ATP synthase—the structure of the stator stalk. Trends Biochem Sci 32:53–56. Essential Cell Biology, Fourth Edition REFERENCES Electron-­‐Transport Chains and Their Proton Pumps Beinert H, Holm RH & Munck E (1997) Iron-­‐sulfur clusters: nature’s modular, multipurpose structures. Science 277:653–659. Berry EA, Guergova-­‐Kuras M, Huang LS & Crofts AR (2000) Structure and function of cytochrome bc complexes. Annu Rev Biochem 69:1005–1075. Brand MD (2005) The efficiency and plasticity of mitochondrial energy transduction. Biochem Soc Trans 33:897–904. Brandt U (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem 75:69–92. Chance B & Williams GR (1955) A method for the localization of sites for oxidative phosphorylation. Nature 176:250–254. Gottschalk G (1997) Bacterial Metabolism, 2nd ed. New York: Springer. Gray HB & Winkler JR (1996) Electron transfer in proteins. Annu Rev Biochem 65:537–556. Keilin D (1966) The History of Cell Respiration and Cytochromes. Cambridge: Cambridge University Press. Sazanov LA (2007) Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46:2275–2288. Subramaniam S & Henderson R (2000) Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature 406:653–657. Tsukihara T, Aoyama H, Yamashita E et al (1996) The whole structure of the 13-­‐subunit oxidized cytochrome c oxidase at 2.8Å. Science 272:1136– 1144. Wikstrom M & Verkhovsky MI (2006) Towards the mechanism of proton pumping by the haem-­‐copper oxidases. Biochim Biophys Acta 1757:1047–1051. Chloroplasts and Photosynthesis Blankenship RE (2002) Molecular Mechanisms of Photosynthesis. Oxford, UK:Blackwell Scientific. Bassham JA (1962) The path of carbon in photosynthesis. Sci Am 206:88–100. Deisenhofer J & Michel H (1989) Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. EMBO J 8:2149–2170. Edwards GE, Furbank RT, Hatch MD & Osmond CB (2001) What does it take to be c(4)?—lessons from the evolution of c(4) photosynthesis. Plant Physiol 125:46–49. Ferreira KN, Iverson TM, Maghlaoui K et al (2004) Architecture of the photosynthetic oxygen-­‐evolving center. Science 303:1831–1838. Iwata S & Barber J (2004) Structure of photosystem II and molecular architecture of the oxygen-­‐evolving centre. Curr Opin Struct Biol 14:447–453. Jordan P, Fromme P, Witt HT et al (2001) Three-­‐ dimensional structure of cyanobacterial photosystem I at 2.5Å resolution. Nature 411:909– 917. Ling Q et al. (2012) Chloroplast biogeneisis is regulated by direct action of the ubiquitin-­‐proteasome system. Science 338(6107): 655-­‐9. Merchant S & Sawaya MR (2005) The light reactions: A guide to recent acquisitions for the picture gallery. The Plant Cell 17:648–663. Munekage Y et al. (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429(6991): 579-­‐82. Nelson N & Ben-­‐Shem A (2004) The complex architecture of oxygenic photosynthesis. Nature Rev Mol Cell Biol 5:971–982. Nevo R et al. (2012) Composition, architecture, and dynamics of the photosynthetic apparatus in higher plants. Plant K 70(1): 157-­‐76. The Evolution of Energy-­‐Generating Systems Blankenship RE & Bauer CE (eds) (1995) Anoxygenic Photosynthetic Bacteria. Dordrecht: Kluwer. de Las Rivas J, Balsera M & Barber J (2004) Evolution of oxygenic photosynthesis: genome-­‐wide analysis of the OEC extrinsic proteins. Trends Plant Sci 9:18–25. Hohmann-­‐Marriott MF, Blankenship RE (2011) Evolution of photosynthesis. Annu Rev Plant Biol 62:515-­‐48. Olson JM, Blankenship RE (2004) Thinking about the evolution of photosynthesis. Photosynth Res 70(1-­‐3): 373-­‐86. Orgel LE (1998) The origin of life—a review of facts and speculations. Trends Biochem Sci. 23:491–495. Pierron D et al. (2012) Evolution of the couple cytochrome c and cytochrome c oxidase in primates. Adv Exp Med Biol 748: 185-­‐213. Schafer G, Purschke W & Schmidt CL (1996) On the origin of respiration: electron transport proteins from archaea to man. FEMS Microbiol Rev 18:173– 188. Skulachev VP (1994) Bioenergetics: the evolution of molecular mechanisms and the development of bioenergetic concepts. Antonie Van Leeuwenhoek 65:271–284. Essential Cell Biology, Fourth Edition REFERENCES Chapter 15 The Compartmentalization of Cells Blobel G (1980) Intracellular protein topogenesis. Proc Natl Acad Sci USA 77:1496–1500. De Duve C (2007) The origin of eukaryotes: a reappraisal. Nature Rev Genet 8:395–403. Schatz G & Dobberstein B (1996) Common principles of protein translocation across membranes. Science 271:1519–1526. Sharma SK, Christen P, Goloubinoff P (2009) Disaggregating chaperones: an unfolding story. Curr Protein Pept Sci 10(5):432-­‐46. Warren G & Wickner W (1996) Organelle inheritance. Cell 84:395–400. The Transport of Molecules Between the Nucleus and the Cytosol Adam SA & Gerace L (1991) Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66:837–847. Adams RL, Wente SR (2013) Uncovering nuclear pore complexity with innovation. Cell 152(6): 1218-­‐21. Bednenko J, Cingolani G & Gerace L (2003) Nucleocytoplasmic transport: navigating the channel. Traffic 4:127–135. Chook YM & Blobel G (2001) Karyopherins and nuclear import. Curr Opin Struct Biol 11:703–715. Cole CN & Scarcelli JJ (2006) Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol 18:299–306. Fahrenkrog B, Koser J & Aebi U (2004) The nuclear pore complex: a jack of all trades? Trends Biochem Sci 29:175–182. Görlich D & Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15:607–660. Hetzer MW, Walther TC & Mattaj IW (2005) Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 21:347–380. Komeili A & O’Shea EK (2000) Nuclear transport and transcription. Curr Opin Cell Biol 12:355–360. Kuersten S, Ohno M & Mattaj IW (2001) Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol 11:497–503. Tran EJ & Wente SR (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125:1041–1053. Weis K (2002) Nucleocytoplasmic transport: cargo trafficking across the border. Curr Opin Cell Biol 14:328–335. The Transport of Proteins Into Mitochondria and Chloroplasts Jarvis P & Robinson C (2004) Mechanisms of protein import and routing in chloroplasts. Curr Biol 14:R1064–R1077. Kessler F & Schnell DJ (2004) Chloroplast protein import: solve the GTPase riddle for entry. Trends Cell Biol 14:334–338. Koehler CM, Merchant S & Schatz G (1999) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem Sci 24:428– 432. Mokranjac D & Neupert W (2005) Protein import into mitochondria. Biochem Soc Trans 33:1019–1023. Prakash S & Matouschek A (2004) Protein unfolding in the cell. Trends Biochem Sci 29:593–600. Soll J & Schleiff E (2004) Protein import into chloroplasts. Nature Rev Mol Cell Biol 5:198–208. Truscott KN, Brandner K & Pfanner N (2003) Mechanisms of protein import into mitochondria. Curr Biol 13:R326–R337. The Endoplasmic Reticulum Adelman MR, Sabatini DD et al (1973) Ribosome-­‐ membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol 56:206–229. Bernales S, Papa FR & Walter P (2006) Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22:487–508. Bishop WR & Bell RM (1988) Assembly of phospholipids into cellular membranes: biosynthesis, transmembrane movement and intracellular translocation. Annu Rev Cell Biol 4:579–610. Blobel G & Dobberstein B (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-­‐bound ribosomes of murine myeloma. J Cell Biol 67:835–851. Borgese N, Mok W & Sabatini DD (1974) Ribosomal-­‐ membrane ineraction: in vitro binding of ribosomes to microsomal membranes. Chen S, Novick P, Ferro-­‐Novick S (2013): ER structure and function. Curr Opin Cell Biol 25(4)428-­‐33. Daleke DL (2003) Regulation of transbilayer plasma membrane phospholipid asymmetry. J Lipid Res 44:233–242. Deshaies RJ, Sanders SL & Schekman R (1991) Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-­‐bound multisubunit complex. Nature 349:806–808. Essential Cell Biology, Fourth Edition REFERENCES Dobson CM (2003) Protein folding and misfolding, Nature 426(6968): 884-­‐90. Ellgaard L & Helenius A (2003) Quality control in the endoplasmic reticulum. Nature Rev Mol Cell Biol 4:181–191. Ferguson MA (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci 112:2799–2809. Gething MJ (1999) Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol 10:465–472. Görlich D, Prehn S et al (1992) A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71:489–503. Helenius J & Aebi M (2002) Transmembrane movement of dolichol linked carbohydrates during N-­‐glycoprotein biosynthesis in the endoplasmic reticulum. Semin Cell Dev Biol 13:171–178. Johnson AE & van Waes MA (1999) The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol 15:799–842. Keenan RJ, Freymann DM & Walter P (2001) The signal recognition particle. Annu Rev Biochem 70:755–775. Kostova Z & Wolf DH (2003) For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-­‐proteasome connection. EMBO J 22:2309–2317. Levine T & Loewen C (2006) Inter-­‐organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol 18:371–378. Marciniak SJ & Ron D (2006) Endoplasmic reticulum stress signaling in disease. Physiol Rev 86:1133– 1149. Milstein C, Brownlee GG et al (1972) A possible precursor of immunoglobulin light chains. Nature New Biol 239:117–120. Römisch K (2005) Endoplasmic reticulum-­‐associated degradation. Annu Rev Cell Dev Biol 21:435–456. Simon SM & Blobel G (1991) A protein-­‐conducting channel in the endoplasmic reticulum. Cell 65:371– 380. Staehelin LA (1997) The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J 11:1151–1165. Trombetta ES & Parodi AJ (2003) Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol 19:649–676. Tsai B, Ye Y & Rapoport TA (2002) Retro-­‐translocation of proteins from the endoplasmic reticulum into the cytosol. Nature Rev Mol Cell Biol 3:246–255. Shibata Y, Voeltz GK & Rapoport TA (2006) Rough sheets and smooth tubules. Cell 126:435–439 White SH & von Heijne G (2004) The machinery of membrane protein assembly. Curr Opin Struct Biol 14:397–404. Yan A & Lennarz WJ (2005) Unraveling the mechanism of protein N-­‐glycosylation. J Biol Chem 280:3121– 3124. Chapter 16 General Principles of Cell Communication Ben-­‐Shlomo I, Yu Hsu S, Rauch R et al (2003) Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Sci STKE 187:RE9. Bourne HR (1995) GTPases: a family of molecular switches and clocks. Philos Trans R Soc Lond B Biol Sci 349:283–289. Ferrell JE Jr (2002) Self-­‐perpetuating states in signal transduction: positive feedback, double-­‐negative feedback and bistability. Curr Opin Cell Biol 14:140– 148. Mangelsdorf DJ, Thummel C, Beato M et al (1995) The nuclear receptor superfamily: the second decade. Cell 83:835–839. Murad F (2006) Shattuck Lecture. Nitric oxide and cyclic GMP in cell signaling and drug development. N Engl J Med 355:2003–2011. Papin JA, Hunter T, Palsson BO & Subramaniam S (2005) Reconstruction of cellular signalling networks and analysis of their properties. Nature Rev Mol Cell Biol 6:99–111. Pawson T & Scott JD (2005) Protein phosphorylation in signaling—50 years and counting. Trends Biochem Sci 30:286–290. Pires-­‐daSilva A & Sommer RJ (2003) The evolution of signalling pathways in animal development. Nature Rev Genet 4:39–49. Robinson-­‐Rechavi M, Escriva Garcia H & Laudet V (2003) The nuclear receptor superfamily. J Cell Sci 116:585–586. Seet BT, Dikic I, Zhou MM & Pawson T (2006) Reading protein modifications with interaction domains. Nature Rev Mol Cell Biol 7:473–483. Singla V & Reiter JF (2006) The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science 313:629–633. Signaling Through G-­‐protein-­‐coupled Cell-­‐surface Receptors Audet M, Bouvier M (2012) Restructuring G-­‐protein coupled receptor activation. Cell 151(1): 14-­‐23. Essential Cell Biology, Fourth Edition REFERENCES Berridge MJ (2005) Unlocking the secrets of cell signaling. Annu Rev Physiol 67:1–21. Berridge MJ, Bootman MD & Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nature Rev Mol Cell Biol 4:517–529. Breer H (2003) Sense of smell: recognition and transduction of olfactory signals. Biochem Soc Trans 31:113–116. Burns ME & Baylor DA (2001) Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu Rev Neurosci 24:779–805. Cooper DM (2005) Compartmentalization of adenylate cyclase and cAMP signalling. Biochem Soc Trans 33:1319–1322. Hoeflich KP & Ikura M (2002) Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108:739–742. Hudmon A & Schulman H (2002) Structure-­‐function of 2+ the multifunctional Ca /calmodulin-­‐dependent protein kinase II. Biochem J 364:593–611. Katritch V, Cherezov V, Stevens RC (2013) Structure-­‐ function of the G-­‐protein coupled receptor superfamily. Annu Rev Pharmacol Toxicol 53: 531-­‐56 Kamenetsky M, Middelhaufe S, Bank EM et al (2006) Molecular details of cAMP generation in mammalian cells: a tale of two systems. J Mol Biol 362:623–639. Latek D et al. (2012) G protein-­‐coupled receptors— recent advances. Acta Biochim Pol 59(4): 515-­‐29 Luttrell LM (2006) Transmembrane signaling by G protein-­‐coupled receptors. Methods Mol Biol 332:3– 49. McConnachie G, Langeberg LK & Scott JD (2006) AKAP signaling complexes: getting to the heart of the matter. Trends Mol Med 12:317–323. Parker PJ (2004) The ubiquitous phosphoinositides. Biochem Soc Trans 32:893–898. Pierce KL, Premont RT & Lefkowitz RJ (2002) Seven-­‐ transmembrane receptors. Nature Rev Mol Cell Biol 3:639–650. Preininger AM, Meiler J, Hamm HE (2013) Conformational flexibility and structural dynamics in GPCR-­‐mediated G protein activation: a perspective. J Mol Biol 425(13): 2288-­‐98. Reiter E & Lefkowitz RJ (2006) GRKs and beta-­‐arrestins: roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab 17:159–165. Rhee SG (2001) Regulation of phosphoinositide-­‐specific phospholipase C. Annu Rev Biochem 70:281–312. Robishaw JD & Berlot CH (2004) Translating G protein subunit diversity into functional specificity. Curr Opin Cell Biol 16:206–209. Shaywitz AJ & Greenberg ME (1999) CREB: a stimulus-­‐ induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 68:821–861. Venkatakrishnan AJ et al. (2013) Molecular signatures of G-­‐protein coupled receptors. Nature 494(7436):185-­‐94 Signaling Through Enzyme-­‐coupled Cell-­‐surface Receptors Baker MD, Wolanin PM & Stock JB (2006) Signal transduction in bacterial chemotaxis. BioEssays 28:9–22. Dard N & Peter M (2006) Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. BioEssays 28:146–156. Darnell JE Jr, Kerr IM & Stark GR (1994) Jak-­‐STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421. Downward J (2004) PI 3-­‐kinase, Akt and cell survival. Semin Cell Dev Biol 15:177–182. Jaffe AB & Hall A (2005) Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 21:247–269. Massague J & Gomis RR (2006) The logic of TGFb signaling. FEBS Lett 580:2811–2820. Mitin N, Rossman KL & Der CJ (2005) Signaling interplay in Ras superfamily function. Curr Biol 15:R563–574. Murai KK & Pasquale EB (2003) Eph’ective signaling: forward, reverse and crosstalk. J Cell Sci 116:2823– 2832. Pawson T (2004) Specificity in signal transduction: from phosphotyrosine-­‐SH2 domain interactions to complex cellular systems. Cell 116:191–203. Qi M & Elion EA (2005) MAP kinase pathways. J Cell Sci 118:3569–3572. Rawlings JS, Rosler KM & Harrison DA (2004) The JAK/STAT signaling pathway. J Cell Sci 117:1281– 1283. Roskoski R Jr (2004) Src protein-­‐tyrosine kinase structure and regulation. Biochem Biophys Res Commun 324:1155–1164. Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103:211–225. Sahin M, Greer PL, Lin MZ et al (2005) Eph-­‐dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron 46:191–204. Schwartz MA & Madhani HD (2004) Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet 38:725–748. Essential Cell Biology, Fourth Edition REFERENCES Shaw RJ & Cantley LC (2006) Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 44:424–430. Wassarman DA, Therrien M & Rubin GM (1995) The Ras signaling pathway in Drosophila. Curr Opin Genet Dev 5:44–50. Wullschleger S, Loewith R & Hall MN (2006) TOR signaling in growth and metabolism. Cell 124:471– 484. Chapter 17 The Self-­‐Assembly and Dynamic Structure of Cytoskeletal Filaments Coulombe PA et al. (1991) A function for keratins and a common thread among different types of epidermolysis bullosa simplex diseases. J Cell Biol 115: 1661-­‐74. Dogterom M & Yurke B (1997) Measurement of the force-­‐velocity relation for growing microtubules. Science 278:856–860. Garner EC, Campbell CS & Mullins RD (2004) Dynamic instability in a DNA-­‐segregating prokaryotic actin homolog. Science 306:1021–1025. Helfand BT, Chang L & Goldman RD (2003) The dynamic and motile properties of intermediate filaments. Annu Rev Cell Dev Biol 19:445–467. Hill TL & Kirschner MW (1982) Bioenergetics and kinetics of microtubule and actin filament assembly-­‐ disassembly. Int Rev Cytol 78:1–125. Hotani H & Horio T (1988) Dynamics of microtubules visualized by darkfield microscopy: treadmilling and dynamic instability. Cell Motil Cytoskeleton 10:229– 236. Jones LJ, Carballido-­‐Lopez R & Errington J (2001) Control of cell shape in bacteria: helical, actin-­‐like filaments in Bacillus subtilis. Cell 104:913–922. Kerssemakers JW, Munteanu EL, Laan L et al (2006) Assembly dynamics of microtubules at molecular resolution. Nature 442:709–712. Luby-­‐Phelps K (2000) Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol 192:189–221. Mitchison T & Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312:237–242. Mitchison TJ (1995) Evolution of a dynamic cytoskeleton. Philos Trans R Soc Lond B Biol Sci 349:299–304. Mukherjee A & Lutkenhaus J (1994) Guanine nucleotide-­‐dependent assembly of FtsZ into filaments. J Bacteriol 176:2754–2758. Oosawa F & Asakura S (1975) Thermodynamics of the polymerization of protein. New York: Academic Press, pp 41–55, pp 90–108. Pauling L (1953) Aggregation of Globular Proteins. Discuss Faraday Soc 13:170–176 Rodionov VI & Borisy GG (1997) Microtubule treadmilling in vivo. Science. 275:215–218. Shih YL & Rothfield L (2006) The bacterial cytoskeleton. Microbiol Mol Biol Rev 70:729–754. Theriot JA (2000) The polymerization motor. Traffic 1:19–28. How Cells Regulate Their Cytoskeletal Filaments Aldaz H, Rice LM, Stearns T & Agard DA (2005) Insights into microtubule nucleation from the crystal structure of human gamma-­‐tubulin. Nature 435:523–527 Bretscher A, Chambers D, Nguyen R & Reczek D (2000) ERM-­‐Merlin and EBP50 protein families in plasma membrane organization and function. Annu Rev Cell Dev Biol 16:113–143. Doxsey S, McCollum D & Theurkauf W (2005) Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 21:411–434. Garcia ML & Cleveland DW (2001) Going new places using an old MAP: tau, microtubules and human neurodegenerative disease. Curr Opin Cell Biol 13:41–48. Holy TE, Dogterom M, Yurke B & Leibler S (1997) Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci USA 94:6228–6231. Mullins RD, Heuser JA & Pollard TD (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA 95:6181–6186. Robinson RC, Turbedsky K, Kaiser DA et al (2001) Crystal structure of Arp2/3 complex. Science 294:1679–1684. Quarmby L (2000) Cellular Samurai: katanin and the severing of microtubules. J Cell Sci 113:2821–2827. Stearns T & Kirschner M (1994) In vitro reconstitution of centrosome assembly and function: the central role of gamma-­‐tubulin. Cell 76:623–637. Wiese C & Zheng Y (2006) Microtubule nucleation: gamma-­‐tubulin and beyond. J Cell Sci 119:4143– 4153. Zheng Y, Wong ML, Alberts B & Mitchison T (1995) Nucleation of microtubule assembly by a gamma-­‐ tubulin-­‐containing ring complex. Nature 378:578– 583. Essential Cell Biology, Fourth Edition REFERENCES Zigmond SH (2004) Formin-­‐induced nucleation of actin filaments. Curr Opin Cell Biol 16:99–105. Molecular Motors Burgess SA, Walker ML, Sakakibara H et al (2003) Dynein structure and power stroke. Nature 421:715– 718. Hirokawa N (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279:519–526. Howard J, Hudspeth AJ & Vale RD (1989) Movement of microtubules by single kinesin molecules. Nature 342:154–158. Howard J (1997) Molecular motors: structural adaptations to cellular functions. Nature 389:561– 567. Kikkawa M (2013) Big steps toward understanding dynein. J Cell Biol 202(1): 15-­‐23. Rayment I, Rypniewski WR, Schmidt-­‐Base K et al (1993) Three-­‐dimensional structure of myosin subfragment-­‐ 1: a molecular motor. Science 261:50–58. Reck-­‐Peterson SL, Yildiz A, Carter AP et al (2006) Single-­‐ molecule analysis of dynein processivity and stepping behavior. Cell 126:335–348. Rice S, Lin AW, Safer D et al (1999) A structural change in the kinesin motor protein that drives motility. Nature 402:778–784. Richards TA & Cavalier-­‐Smith T (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113–1118. Svoboda K, Schmidt CF, Schnapp BJ & Block SM (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365:721–727. Vikstrom KL & Leinwand LA (1996) Contractile protein mutations and heart disease. Curr Opin Cell Biol 8:97–105. Wells AL, Lin AW, Chen LQ et al (1999) Myosin VI is an actin-­‐based motor that moves backwards. Nature 401:505–508. Yildiz A, Forkey JN, McKinney SA et al (2003) Myosin V walks hand-­‐overhand: single fluorophore imaging with 1.5-­‐nm localization. Science 300:2061–2065. Yildiz A & Selvin PR (2005) Kinesin: walking, crawling or sliding along? Trends Cell Biol 15:112–120. The Cytoskeleton and Cell Behavior Abercrombie M (1980) The crawling movement of metazoan cells. Proc Roy Soc B 207:129–147. Cooke R (2004) The sliding filament model: 1972–2004. J Gen Physiol 123:643–656. Cooper JA (2013) Cell biology in neuroscience: mechanisms of cell migration in the nervous system. Cell Biol 202(5): 725-­‐34. Dent EW & Gertler FB (2003) Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40:209–227. Kikkawa M (2013) Big steps toward understanding dynein. J Cell Biol 202(1) 15-­‐23. Lauffenburger DA & Horwitz AF (1996) Cell migration: a physically integrated molecular process. Cell 84:359– 369. Lo CM, Wang HB, Dembo M & Wang YL (2000) Cell movement is guided by the rigidity of the substrate. Biophys J 79:144–152. Madden K & Snyder M (1998) Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol 52:687–744. Ridley AJ, Schwartz MA, Burridge K et al (2003) Cell migration: integrating signals from front to back. Science 302:1704–1709. Rafelski SM & Theriot JA (2004) Crawling toward a unified model of cell motility: spatial and temporal regulation of actin dynamics. Annu Rev Biochem 73:209–239. Parent CA & Devreotes PN (1999) A cell’s sense of direction. Science 284:765–770. Pollard TD & Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–465. Purcell EM (1977) Life at low Reynolds’ number. Am J Phys 45:3–11. Shirao T, Gonzalez-­‐Billaut C (2013) Actin filaments and microtubules in dendritic spines. J Neurochem 126(2): 155-­‐64. Wittmann T, Hyman A & Desai A (2001) The spindle: a dynamic assembly of microtubules and motors. Nature Cell Biol 3:E28–E34. Chapter 18 Overview of the Cell Cycle Forsburg SL & Nurse P (1991) Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu Rev Cell Biol 7:227–256. Hartwell LH, Culotti J, Pringle JR et al (1974) Genetic control of the cell division cycle in yeast. Science 183:46–51. Kirschner M, Newport J & Gerhart J (1985) The timing of early developmental events in Xenopus. Trends Genet 1:41–47. Nurse P, Thuriaux P & Nasmyth K (1976) Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 146:167–178. Essential Cell Biology, Fourth Edition REFERENCES The Cell-­‐Cycle Control System Evans T, Rosenthal ET, Youngblom J et al (1983) Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33:389–396. Lohka MJ, Hayes MK & Maller JL (1988) Purification of maturation-­‐promoting factor, an intracellular regulator of early mitotic events. Proc Natl Acad Sci USA 85:3009–3013. Masui Y and Markert CL (1971) Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool 177:129–146. Morgan DO (1997) Cyclin-­‐dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13:261–291. Murray AW & Kirschner MW (1989) Cyclin synthesis drives the early embryonic cell cycle. Nature 339:275–280. Pavletich NP (1999) Mechanisms of cyclin-­‐dependent kinase regulation: structures of Cdks, their cyclin activators, and CIP and Ink4 inhibitors. J Mol Biol 287:821–828. Peters JM (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nature Rev Mol Cell Biol 7:644–656. Petroski MD & Deshaies RJ (2005) Function and regulation of cullin-­‐RING ubiquitin ligases. Nature Rev Mol Cell Biol 6:9–20. Wittenberg C & Reed SI (2005) Cell cycle-­‐dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24:2746– 2755. S Phase Arias EE & Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21:497–518. Bell SP & Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71:333–374. Bell SP & Stillman B (1992) ATP-­‐dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128–134. Diffley JF (2004) Regulation of early events in chromosome replication. Curr Biol 14:R778–R786. Groth A, Rocha W, Verreault A et al (2007) Chromatin challenges during DNA replication and repair. Cell 128:721–733. Tanaka S, Umemori T, Hirai K et al (2007) CDK-­‐ dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445:328– 332. Zegerman P & Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-­‐dependent kinases promotes DNA replication in budding yeast. Nature 445:281–285. Mitosis Cheeseman IM, Chappie JS, Wilson-­‐Kubalek EM et al (2006) The conserved KMN network constitutes the core microtubule-­‐binding site of the kinetochore. Cell 127:983–997. Dong Y, Vanden Beldt KJ, Meng X et al (2007) The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nature Cell Biol 9:516–522. Heald R, Tournebize R, Blank T et al (1996) Self-­‐ organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382:420–425. Hirano T (2005) Condensins: organizing and segregating the genome. Curr Biol 15:R265–R275. Kapoor TM, Lampson MA, Hergert P et al (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311:388–391. Mitchison T & Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312:237–242. Mitchison TJ (1989) Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol 109:637–652. Mitchison TJ & Salmon ED (2001) Mitosis: a history of division. Nature Cell Biol 3:E17–E21. Musacchio A & Salmon ED (2007) The spindle-­‐assembly checkpoint in space and time. Nature Rev Mol Cell Biol 8:379–393. Nasmyth K (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559–565. Nigg EA (2007) Centrosome duplication: of rules and licenses. Trends Cell Biol 17:215–221. Nurse P (1990) Universal control mechanism regulating onset of M-­‐phase. Nature 344:503–508. Page SL & Hawley RS (2003) Chromosome choreography: the meiotic ballet. Science 301:785– 789. Petronczki M, Siomos MF & Nasmyth K (2003) Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423– 440. Salmon ED (2005) Microtubules: a ring for the depolymerization motor. Curr Biol 15:R299–R302. Tanaka TU, Stark MJ & Tanaka K (2005) Kinetochore capture and biorientation on the mitotic spindle. Nature Rev Mol Cell Biol 6:929–942. Uhlmann F, Lottspeich F & Nasmyth K (1999) Sister-­‐ chromatid separation at anaphase onset is promoted Essential Cell Biology, Fourth Edition REFERENCES by cleavage of the cohesin subunit Scc1. Nature 400:37–42. Wadsworth P & Khodjakov A (2004) E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol 14:413–419. Cytokinesis Albertson R, Riggs B & Sullivan W (2005) Membrane traffic: a driving force in cytokinesis. Trends Cell Biol 15:92–101. Burgess DR & Chang F (2005) Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 15:156–162. Dechant R & Glotzer M (2003) Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev Cell 4:333– 344. Eggert US, Mitchison TJ & Field CM (2006) Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem 75:543–566. Glotzer M (2005) The molecular requirements for cytokinesis. Science 307:1735–1739. Grill SW, Gonczy P, Stelzer EH et al (2001) Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409:630–633. Jurgens G (2005) Plant cytokinesis: fission by fusion. Trends Cell Biol 15:277–283. Rappaport R (1986) Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol 105:245– 281. Control of Cell Division and Cell Growth Adhikary S & Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nature Rev Mol Cell Biol 6:635–645. Campisi J (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120:513–522. Conlon I & Raff M (1999) Size control in animal development. Cell 96:235–244. Frolov MV, Huen DS, Stevaux O et al (2001) Functional antagonism between E2F family members. Genes Dev 15:2146–2160. Harrison JC & Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40:209–235. Jorgensen P & Tyers M (2004) How cells coordinate growth and division. Curr Biol 14:R1014–R1027. Levine AJ (1997) p53, the cellular gatekeeper for growth and division. Cell 88:323–331. Lee SJ (2007) Quadrupling muscle mass in mice by targeting TGF-­‐beta signaling pathways. PLoS One 2:e789. Raff MC (1992) Social controls on cell survival and cell death. Nature 356:397–400. Sherr CJ & DePinho RA (2000) Cellular senescence: mitotic clock or culture shock? Cell 102:407–410. Sherr CJ & Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-­‐phase progression. Genes Dev 13:1501–1512. Trimarchi JM & Lees JA (2002) Sibling rivalry in the E2F family. Nature Rev Mol Cell Biol 3:11–20. Vousden KH & Lu X (2002) Live or let die: the cell’s response to p53. Nature Rev Cancer 2:594–604. Zetterberg A & Larsson O (1985) Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc Natl Acad Sci USA 82:5365–5369. Apoptosis Adams JM, Huang DC, Strasser A et al (2005) Subversion of the Bcl-­‐2 life/death switch in cancer development and therapy. Cold Spring Harb Symp Quant Biol 70:469–77. Boatright KM & Salvesen GS (2003) Mechanisms of caspase activation. Curr Opin Cell Biol 15:725–731. Danial NN & Korsmeyer SJ (2004) Cell death: critical control points. Cell 116:205–219. Ellis RE, Yuan JY & Horvitz RA (1991) Mechanisms and functions of cell death. Annu Rev Cell Biol 7:663–698. Fadok VA & Henson PM (2003) Apoptosis: giving phosphatidylserine recognition an assist—with a twist. Curr Biol 13:R655–R657. Galonek HL & Hardwick JM (2006) Upgrading the BCL-­‐2 network. Nature Cell Biol 8:1317–1319. Green DR (2005) Apoptotic pathways: ten minutes to dead. Cell 121:671–674. Horvitz HR (2003) Worms, life, and death (Nobel lecture). Chembiochem 4:697–711. Hyun-­‐Eui K, Fenghe D, Fang M & Wang X (2005) Formation of apoptosome is initiated by cytochrome c-­‐induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-­‐1. Proc Natl Acad Sci USA 102:17545–17550. Jacobson MD, Weil M & Raff MC (1997) Programmed cell death in animal development. Cell 88:347–354. Jiang X & Wang X (2004) Cytochrome C-­‐mediated apoptosis. Annu Rev Biochem 73:87–106. Kerr JF, Wyllie AH & Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-­‐ranging implications in tissue kinetics. B J Cancer 26:239– 257. Essential Cell Biology, Fourth Edition REFERENCES Kumar S (2007) Caspase function in programmed cell death. Cell Death Differ 14:32–43. Lavrik I, Golks A & Krammer PH (2005) Death receptor signaling. J Cell Sci 118:265–267. Lowe SW, Cepero E & Evan G (2004) Intrinsic tumour suppression. Nature 432:307–315. Lum JJ, Bauer DE, Kong M et al (2005) Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120:237–48. McCall K & Steller H (1997) Facing death in the fly: genetic analysis of apoptosis in Drosophila. Trends Genet 13:222–226. Mills Kd (2013) Tumor suppression: Putting p53 in context. Cell Cyce 12(22). Nagata S (1999) Fas ligand-­‐induced apoptosis. Annu Rev Genet 33:29–55. Nagata S (2005) DNA degradation in development and programmed cell death. Annu Rev Immunol 23:853– 875. Pop C, Timmer J, Sperandio S & Salvesen GS (2006) The apoptosome activates caspase-­‐9 by dimerization. Mol Cell 22:269–275. Raff MC (1999) Cell suicide for beginners. Nature 396:119–122. Rathmell JC & Thompson CB (2002) Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell 109:S97–107. Tittel JN & Steller H (2000) A comparison of programmed cell death between species. Genome Biol 1. Verhagen AM, Coulson EJ & Vaux DL (2001) Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol 2:3009.1–3009.10. Vousden KH (2005) Apoptosis. p53 and PUMA: a deadly duo. Science 309:1685–1686. Willis SN & Adams JM (2005) Life in the balance: how BH3-­‐only proteins induce apoptosis. Curr Opin Cell Biol 17:617–625. Chapter 19 Overview of Sexual Reproduction Cavalier-­‐Smith T (2002) Origins of the machinery of recombination and sex. Heredity 88:125–41. Charlesworth B (2006) The evolutionary biology of sex. Curr Biol 16:R693–R695. Hoekstra RF (2005) Evolutionary biology: why sex is good. Nature 434:571–573. Maynard Smith J (1978) Evolution of Sex. Cambridge, UK: Cambridge University Press. Meiosis Blat Y, Protacio RU, Hunter N and Kleckner N (2002) Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111:791– 802. Börner GV, Kleckner N and Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117:29–45. De Massy B (2003) Distribution of meiotic recombination sites. Trends Genet 19:514–522. Gerton JL, and Hawley RS (2005) Homologous chromosome interactions in meiosis: diversity amidst conservation. Nature Rev Genet 6:477–487. Hall H, Hunt P and Hassold T (2006) Meiosis and sex chromosome aneuploidy: how meiotic errors cause aneuploidy; how aneuploidy causes meiotic errors. Curr Opin Genet Dev 16:323–329. Hauf S and Watanabe Y (2004) Kinetochore orientation in mitosis and meiosis. Cell 119:317–327. Hunt PA and Hassold TJ (2002) Sex matters in meiosis. Science 296:2181–2183. Jordan P (2006) Initiation of homologous chromosome pairing during meiosis. Biochem Soc Trans 34:545– 549. Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35:673–745. Page SL and Hawley RS (2004) The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20:525–558. Petronczki M, Siomos MF and Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423– 440. Fertilization De Jonge C (2005) Biological basis for human capacitation. Hum Reprod Update 11:205–214. Hafez ES, Goff L and Hafez B (2004) Mammalian fertilization, IVF, ICSI:physiological/molecular parameters, clinical application. Arch Androl 50:69– 88. Jaenisch R (2004) Human Cloning—The Science and Ethics of Nuclear Transplantation. N Eng J Med 351:2787–2791. Shur BD, Rodeheffer C and Ensslin MA (2004) Mammalian fertilization. Curr Biol 14:R691–692. Stein KK, Primakoff P and Myles D (2004) Sperm-­‐egg fusion: events at the plasma membrane. J Cell Sci 117:6269–6274. Essential Cell Biology, Fourth Edition REFERENCES Takahashi K and Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. Tsaadon A, Eliyahu E, Shtraizent N and Shalgi R (2006) When a sperm meets an egg: block to polyspermy. Mol Cell Endocrinol 252:107–114. Wassarman PM (2005) Contribution of mouse egg zona pellucida glycoproteins to gamete recognition during fertilization. J Cell Physiol 204:388–391. Whitaker M (2006) Calcium at fertilization and in early development. Physiol Rev 86:25–88. Inheritence and Genetics Lehner B et al. (2006) Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 38: 896-­‐903. Mardis ER (2013) Next-­‐generation sequencing platofrms. Annu Rev Anal Chem (Palo Alto Calif) 6:287-­‐303. Pareek CS, Smoczynski R, Tretyn A (2011) Sequencing technologies and genome sequencing. J Appl Genet 52(4): 413-­‐35. Chapter 20 Epithelial Sheets and Cell Junctions Furuse M, Hata M, Furuse K et al (2002) Claudin-­‐based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-­‐1-­‐deficient mice. J Cell Biol 156:1099–1111. Ikenouchi J, Furuse M, Furuse K et al (2005) Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171:939–945. Henrique D & Schweisguth F (2003) Cell polarity: the ups and downs of the Par6/aPKC complex. Curr Opin Genet Dev 13:341–350. Nelson WJ (2003) Adaptation of core mechanisms to generate cell polarity. Nature 422:766–774. O’Brien LE, Jou TS, Pollack AL et al (2001) Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nature Cell Biol 3:831–838. Seifert JR & Mlodzik M (2007) Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nature Rev Genet 8:126–138. Shin K, Fogg VC & Margolis B (2006) Tight junctions and cell polarity. Annu Rev Cell Dev Biol 22:207–236. nd Wheater PR et al. (1987) Functional Histology, 2 Edition. London: Churchill Livingstone. Passageways from Cell to Cell: Gap Junctions and Plasmodesmata Cilia ML & Jackson D (2004) Plasmodesmata form and function. Curr Opin Cell Biol 16:500–506. Gaietta G, Deerinck TJ, Adams SR et al (2002) Multicolor and electron microscopic imaging of connexin trafficking. Science 296:503–507. Goodenough DA & Paul DL (2003) Beyond the gap: functions of unpaired connexon channels. Nature Rev Mol Cell Biol 4:285–294. Segretain D & Falk MM (2004) Regulation of connexin biosynthesis, assembly, gap junction formation, and removal. Biochim Biophys Acta 1662:3–21. Wei CJ, Xu X & Lo CW (2004) Connexins and cell signaling in development and disease. Annu Rev Cell Dev Biol 20:811–838. The Basal Lamina Miner JH & Yurchenco PD (2004) Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol 20:255–284. Sanes JR. (2003) The basement membrane/basal lamina of skeletal muscle. J Biol Chem 278:12601– 12604. Sasaki T, Fassler R & Hohenester E (2004) Laminin: the crux of basement membrane assembly. J Cell Biol 164:959–963. Yurchenco PD, Amenta PS & Patton BL (2004) Basement membrane assembly, stability and activities observed through a developmental lens. Matrix Biol 22:521–538. The Extracellular Matrix Aszodi A, Legate KR, Nakchbandi I & Fassler R (2006) What mouse mutants teach us about extracellular matrix function. Annu Rev Cell Dev Biol 22:591–621. Bulow HE & Hobert O (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol 22:375–407. Couchman JR (2003) Syndecans: proteoglycan regulators of cell-­‐surface microdomains? Nature Rev Mol Cell Biol 4:926–937. Hotary KB, Allen ED, Brooks PC et al (2003) Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-­‐dimensional extracellular matrix. Cell 114:33–45. Kielty CM, Sherratt MJ & Shuttleworth CA (2002) Elastic fibres. J Cell Sci 115:2817–2828. Larsen M, Artym VV, Green JA & Yamada KM (2006) The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol 18:463–471. Essential Cell Biology, Fourth Edition REFERENCES Lin X (2004) Functions of heparan sulfate proteoglycans in cell signaling during development. Development 131:6009–6021. Mott JD & Werb Z (2004) Regulation of matrix biology by matrix metalloproteases. Curr Opin Cell Biol 16:558–564. Prockop DJ & Kivirikko KI (1995) Collagens: molecular biology, diseases, and potentials for therapy. Annu Rev Biochem 64:403–434. Toole BP (2001) Hyaluronan in morphogenesis. Semin Cell Dev Biol 12:79–87. The Plant Cell Wall Carpita NC & McCann M (2000) The Cell Wall. In Biochemistry and Molecular Biology of Plants (Buchanan BB,Gruissem W & Jones RL eds), pp 52– 108. Rockville, MD: ASPB. Carpita NC, Campbell M & Tierney M (eds) (2001) Plant cell walls. Plant Mol Biol 47:1–340. (special issue) Dhugga KS (2001) Building the wall: genes and enzyme complexes for polysaccharide synthases. Curr Opin Plant Biol 4:488–493. McCann MC & Roberts K (1991) Architecture of the primary cell wall. In The Cytoskeletal Basis of Plant Growth and Form (Lloyd CW ed), pp 109–129. London: Academic Press. Smith LG & Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21:271–295. Somerville C (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22:53–78. Epidermis and Its Renewal by Stem Cells Fuchs E (2007) Scratching the surface of skin development. Nature 445:834–842. Imagawa W, Yang J, Guzman R & Nandi S (1994) Control of mammary gland development. In The Physiology of Reproduction (Knobil E & Neill JD eds), 2nd ed, pp 1033–1063. New York: Raven Press. Ito M, Yang Z, Andl T et al (2007) Wnt-­‐dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447:316–320. Jacinto A, Martinez-­‐Arias A & Martin P (2001) Mechanisms of epithelial fusion and repair. Nature Cell Biol 3:E117–123. Jensen UB, Lowell S & Watt FM (1999) The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: a new view based on whole-­‐mount labelling and lineage analysis. Development 126:2409–2418. Prince JM, Klinowska TC, Marshman E et al (2002) Cell-­‐ matrix interactions during development and apoptosis of the mouse mammary gland in vivo. Dev Dyn 223:497–516. Shackleton M, Vaillant F, Simpson KJ et al (2006) Generation of a functional mammary gland from a single stem cell. Nature 439:84–88. Shinin V, Gayraud-­‐Morel B, Gomes D & Tajbakhsh S (2006) Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biol 8:677–687. Stanger BZ, Tanaka AJ & Melton DA (2007) Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature 445:886–891. Steinert PM (2000) The complexity and redundancy of epithelial barrier function. J Cell Biol 151:F5–F8. Watt FM, Lo Celso C & Silva-­‐Vargas V (2006) Epidermal stem cells: an update. Curr Opin Genet Dev 16:518– 524. Tissue Maintenance and Renewal Allsopp RC, Morin GB, DePinho R, Harley CB & Weissman IL (2003) Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood 102:517–520. Calvi LM, Adams GB, Weibrecht KW et al (2003) Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841–846. Hock H, Hamblen MJ, Rooke HM et al (2004) Gfi-­‐1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431:1002–1007. Metcalf D (1980) Clonal analysis of proliferation and differentiation of paired daughter cells: action of granulocyte-­‐macrophage colony-­‐stimulating factor on granulocyte-­‐macrophage precursors. Proc Natl Acad Sci USA 77:5327–5330. Metcalf D (1999) Stem cells, pre-­‐progenitor cells and lineage-­‐committed cells: are our dogmas correct? Annu NY Acad Sci 872:289–303. Orkin SH (2000) Diversification of haematopoietic stem cells to specific lineages. Nature Rev Genet 1:57–64. Reya T, Duncan AW, Ailles L et al (2003) A role for Wnt signalling in self-­‐renewal of haematopoietic stem cells. Nature 423:409–414. Shizuru JA, Negrin RS & Weissman IL (2005) Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annu Rev Med 56:509–538. Wintrobe MM (1980) Blood, Pure and Eloquent. New York: McGraw-­‐Hill. Essential Cell Biology, Fourth Edition REFERENCES Stem-­‐cell Engineering Brockes JP & Kumar A (2005) Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science 310:1919–1923. Brustle O, Jones KN, Learish RD et al (1999) Embryonic stem cell-­‐derived glial precursors: a source of myelinating transplants. Science 285:754–756. Conti L, Pollard SM, Gorba T et al (2005) Niche-­‐ independent symmetrical self-­‐renewal of a mammalian tissue stem cell. PLoS Biol 3:e283. Eggan K, Baldwin K, Tackett M et al (2004) Mice cloned from olfactory sensory neurons. Nature 428:44–49. Eiraku M & Sasi Y (2012) Self-­‐formation of layerd neural structures in three-­‐dimensional culture of ES cells. Curr Opin Neurobiol 22:768-­‐777. Eiraku M et al. (2011) Self-­‐forming optic cup morphogenesis in three-­‐dimensional culture. Nature 472:51-­‐56. Lee TI, Jenner RG, Boyer LA et al (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301–313. Lenger CJ (2010) iPS cell technology in regenerative medicine Ann N Y Acad Sci 1192: 38-­‐44. Ming GL & Song H (2005) Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci 28:223–250. Okano H et al (2013) Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res 112)3): 523-­‐33. Okita K, Ichisaka T & Yamanaka S (2007) Generation of germlinecompetent induced pluripotent stem cells. Nature in press. Raff M (2003) Adult stem cell plasticity: fact or artifact? Annu Rev Cell Dev Biol 19:1–22. Schulz JT, 3rd,Tompkins RG & Burke JF (2000) Artificial skin. Annu Rev Med 51:231–244. Suhonen JO, Peterson DA, Ray J & Gage FH (1996) Differentiation of adult hippocampus-­‐derived progenitors into olfactory neurons in vivo. Nature 383:624–627. Tachibana M et al (2013) Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153(6): 1228-­‐38 Wagers AJ & Weissman IL (2004) Plasticity of adult stem cells. Cell 116:639–648. Yu J et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858): 1917-­‐20 Cancer as a Microevolutionary Process Al-­‐Hajj M, Becker MW, Wicha M et al (2004) Therapeutic implications of cancer stem cells. Curr Opin Genet Dev 14:43–47. Bielas JH, Loeb KR, Rubin BP et al (2006) Human cancers express a mutator phenotype. Proc Natl Acad Sci USA 103:18238–18242. Cairns J (1975) Mutation, selection and the natural history of cancer. Nature 255:197–200. Campesi J (2005) Senescent cells, tumor supression, and organismal aging: good citizens, bad neighbors. Cell 120:513–522. Evan GI & Vousden KH (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411:342–348. Fialkow PJ (1976) Clonal origin of human tumors. Biochim Biophys Acta 458:283–321. Fidler IJ (2003) The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature Rev Cancer 3:453–458. Jones PA & Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nature Rev Genet 3:415–428. Loeb LA, Loeb KR & Anderson JP (2003) Multiple mutations and cancer. Proc Natl Acad Sci USA 100:776–781. Lowe SW, Cepero E & Evan G (2004) Intrinsic tumour suppression. Nature 432:307–315. Nowell PC (1976) The clonal evolution of tumor cell populations. Science 194:23–28. Pardal R, Clarke MF & Morrison SJ (2003) Applying the principles of stem-­‐cell biology to cancer. Nature Rev Cancer 3:895–902. Sjoblom T, Jones S, Wood LD et al (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274. Thiery JP (2002) Epithelial-­‐mesenchymal transitions in tumour progression. Nature Rev Cancer 2:442–454. Tlsty TD & Coussens LM (2006) Tumor Stroma and Regulation of Cancer Development. Annu Rev Pathol Mech Dis 1:119–150. Ward RJ & Dirks PB (2007) Cancer stem cells: at the headwaters of tumor development. Annu Rev Pathol Mech Dis 2:175–189. The Preventable Causes of Cancer Ames B, Durston WE, Yamasaki E & Lee FD (1973) Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci USA 70:2281–2285. Berenblum I (1954) A speculative review: the probable nature of promoting action and its significance in the understanding of the mechanism of carcinogenesis. Cancer Res 14:471–477. Cairns J (1985) The treatment of diseases and the war against cancer. Sci Am 253:51–59. Essential Cell Biology, Fourth Edition REFERENCES Doll R (1977) Strategy for detection of cancer hazards to man. Nature 265:589–597. Doll R & Peto R (1981) The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst 66:1191– 1308. Hsu IC, Metcalf RA, Sun T et al (1991) Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 350:427–428. Newton R, Beral V & Weiss RA (eds) (1999) Infections and Human Cancer. Cold Spring Harbor Laboratory Press: Cold Spring Harbor. Peto J (2001) Cancer epidemiology in the last century and the next decade. Nature 411:390–395. Finding the Cancer-­‐Critical Genes Davies H, Bignell GR, Cox C et al (2002) Mutations of the BRAF gene in human cancer. Nature 417:949– 954. Easton DF, Pooley KA, Dunning AM et al (2007) Genome-­‐wide association study identifies novel breast cancer susceptibility loci. Nature 447:1087– 1095. Feinberg AP (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440. Herman JG & Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042–2054. Lowe SW, Cepero E & Evan G (2004) Intrinsic tumour suppression. Nature 432:307–315. Mitelman F, Johansson B & Mertens F (2007) The impact of translocations and gene fusions on cancer causation. Nature Rev Cancer 7:233-­‐245. Pinkel D & Albertson DG (2005). Comparative genomic hybridization. Annu Rev Genomics Hum Genet 6:331–354. Rowley JD (2001) Chromosome translocations: dangerous liasisons revisited. Nature Rev Cancer 1:245–250. The Wellcome Trust Case Control Consortium (2007) Genome-­‐wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–683. Vicente-­‐Duenas C et al. (2013) Function of oncogenes in cancer development: a changing paradigm EMBO J 32(11): 1502-­‐13. Westbrook TF, Stegmeier F & Elledge SJ (2005) Dissecting cancer pathways and vulnerabilities with RNAi. Cold Spring Harb Symp Quant Biol 70:435–444. The Molecular Basis of Cancer Cell Behavior Artandi SE & DePinho RA (2000) Mice without telomerase: what can they teach us about human cancer? Nature Med 6:852–855. Brown JM & Attardi LD (2005) The role of apoptosis in cancer development and treatment response. Nature Rev Cancer 5:231–237. Chambers AF, Naumov GN, Vantyghem S & Tuck AB (2000) Molecular biology of breast cancer metastasis: clinical implications of experimental studies on metastatic inefficiency. Breast Cancer Res 2:400–407. Edwards PAW (1999) The impact of developmental biology on cancer research: an overview. Cancer Metastasis Rev 18:175–180. Futreal PA, Coin L, Marshall M et al (2004) A census of human cancer genes. Nature Rev Cancer 4:177–183. Hanahan D & Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70. Hoffman RM (2005) The multiple uses of fluorescent proteins to visualize cancer in vivo. Nature Rev Cancer 5:796–806. Kinzler KW & Vogelstein B (1996) Lessons from hereditary colorectal cancer. Cell 87:159–170. Levine AJ, Hu W & Feng Z (2006) The P53 pathway: what questions remain to be explored? Cell Death Differ 13:1027–1036. Macleod KF & Jacks T (1999) Insights into cancer from transgenic mouse models. J Pathol 187:43–60. Mani SA, Yang J, Brooks M et al (2007) Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-­‐like breast cancers. Proc Natl Acad Sci USA 104:10069–10074. Mueller MM & Fusenig NE (2004) Friends or foes— bipolar effects of the tumour stroma in cancer. Nature Rev Cancer 4:839–849. Plas DR & Thompson CB (2005) Akt-­‐dependent transformation: there is more to growth than just surviving. Oncogene 24:7435–7442. Ridley A (2000) Molecular switches in metastasis. Nature 406:466–467. Shaw RJ & Cantley LC (2006) Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441:424–430. Sherr CJ (2006) Divorcing ARF and p53: an unsettled case. Nature Rev Cancer 6:663–673. Vogelstein B & Kinzler KW (2004) Cancer genes and the pathways they control. Nature Med 10:789–799. Weinberg RA (1995) The retinoblastoma protein and cell-­‐cycle control. Cell 81:323–330. Essential Cell Biology, Fourth Edition REFERENCES Cancer Treatment: Present and Future Alizadeh AA, Eisen MB, Davis RE et al (2000). Distinct types of diffuse large B-­‐cell lymphoma identified by gene expression profiling. Nature 403:503–511. Baselga J & Albanell J (2001) Mechanism of action of anti-­‐HER2 monoclonal antibodies. Annu Oncol 12:S35–S41. Druker BJ & Lydon NB (2000) Lessons learned from the development of an abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest 105:3– 7. Folkman J (1996) Fighting cancer by attacking its blood supply. Sci Am 275:150–154. Golub TR, Slonim DK, Tamayo P et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537. Huang P & Oliff A (2001) Signaling pathways in apoptosis as potential targets for cancer therapy. Trends Cell Biol 11:343–348. Garraway LA & Sellers WR (2006) Lineage dependency and lineage-­‐survival oncogenes in human cancer. Nature Rev Cancer 6:593–602. Jonkers J & Berns A (2004) Oncogene addiction: sometimes a temporary slavery. Cancer Cell 6:535– 538. Jain RK (2005) Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science 307:58–62. Kim R, Emi M & Tanabe K (2007) Cancer immunoediting from immune surveillance to immune escape. Immunology 121:1–14. Madhusudan S & Middleton MR (2005) The emerging role of DNA repair proteins as predictive, prognostic and therapeutic targets in cancer. Cancer Treat Rev 31:603–617. Roninson IB, Abelson HT, Housman DE et al (1984) Amplification of specific DNA sequences correlates with multi-­‐drug resistance in Chinese hamster cells. Nature 309:626–628. Sarkadi B, Homolya L, Szakács G & Váradi A (2006) Human multidrug resistance ABCB and ABCG transporters: participation in a chemoimmunity defense system. Physiol Rev 86:1179–236. Sawyers C (2004) Targeted cancer therapy. Nature 432:294–297. van ‘t Veer LJ, Dai H, van de Vijver MJ et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536. Varmus H, Pao W, Politi K et al (2005) Oncogenes come of age. Cold Spring Harb Symp Quant Biol 70:1–9. Ventura A, Kirsch DG, McLaughlin ME et al (2007) Restoration of p53 function leads to tumour regression in vivo. Nature 445:661–665.