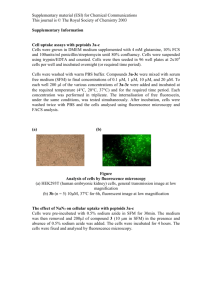
Methods in Cell Biology 2015
advertisement

Methods in Cell Biology 2015 Johannes A. Schmid Internet: www.meduniwien.ac.at/user/johannes.schmid some contents contributed by: Dr. Lukas Mach Institut für Angewandte Genetik und Zellbiologie , Univ. f. Bodenkultur 2 Overview of Topics • • • • • • • • • scientific strategies and assay systems cell culture labelling and transfection of cells analysis of cellular components analysis of molecular interactions fluorescence measurements microscopy flow analysis (fluorescence activated cell sorting, FACS) analysis of various cellular processes (proliferation, apoptosis..) 3 1 Details of the lecture 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. Scientific strategies cell culture labelling and transfection of cells a) radioactive and chemical labelling b) transfections: overexpression of genes and gene suppression c) reporter gene assays gene-suppression (siRNA-technologies) analysis of DNA and proteins (electrophoresis and blotting) subcellular fractionation (centrifugations…) methods to detect macromolecular interactions a) Yeast 1- und 2-hybrid systems b) co-immunoprecipitations c) fluorescence resonance energy transfer (FRET) methods of fluorescence measurements realtime-PCR transmitted light microscopy and contrast principles fluorescence microscopy confocal laser scanning microscopy flow analysis (FACS) analysis of various cellular processes (proliferation, apoptosis..) methods to investigate vesicular transport processes 4 Nodes of regulation in cellular systems cell membrane ligands receptors oligomerization transport signal transduction signal transduction modified protein Golgi transport transcription factor activation posttranslational modification ER protein transport nucleus poly-ubiquitination degradation translation transcription splicing pre-mRNA mRNA mRNA transport DNA micro-RNA processing pre-micro-RNA 5 micro-RNA degradation 2 General scientific strategies • descriptive strategies: the whole system (cell, organism) is observed without influencing it advantage: physiological states are not altered disadvantage: it is difficult to elucidate cause-effect relationships • mechanistic (manipulating) strategies: various factors are kept constant, while others are altered on purpose – the change in the whole system is monitored. advantage : cause-effect relationships can be monitored or detected disadvantage : the physiological steady state is altered and influenced. Results might be artefacts of the measurement system. 6 Experimental Systems I • In vitro, biochemical systems: investigation in a solution (e.g. enzyme reactions): many parameters can be fixed (temperature, pH, buffer composition…). Example: enzyme activity assay, EMSA … • In vitro, cell biological systems: including cellular structures e.g. in vitro-transcription/translation with membrane components, in vitro-fusion of endosomes, nuclear import assays with isolated nuclei etc. Experimental conditions have to be set in a way that cellular structures are not damaged (e.g. isotonic buffer, physiological pH….); reaction partners can be influenced widely (e.g. antibodies can be added…) 7 3 Experimental Systems II • cell culture systems - immortalized cell lines: unlimited passages - primary cells: limited passage number, more complicated cell culture (e.g. coated plates…) - co-culture of different cell types: e.g. keratinocytes + fibroblasts in a collagen matrix. • xenograft systems: - cells are injected into immuno-compromized mice (nude mice, SCID mice); e.g. subcutaneously - tissue recombination systems (e.g. prostate epithelial cells with mesenchymal cells injected into the renal capsule of SCID mice) 8 Experimental Systems III • Organ cultures (e.g. skin sheets, brain slices…) • Organ perfusions 9 4 Experimental Systems IV • Animal Experiments: Just the whole organism provides the full complex biological system that is relevant for most biomedical research topics. The whole organism includes superordinated systems such as the nervous system, the blood circulation, the endocrine system and so on. The cells are in their normal organ environment; cellular communications are intact…. – thus the highest possible physiological state can be achieved. However, specific components of the system (e.g. certain cells) are not easily accessible – and specific experimental manipulations (e.g. of specific cells without side effects) are often difficult). Cause-effect relationships are often difficult to elucidate – and there is a big „black box“ due to the complexity of the system. It has to be considered that results obtained with animals such as mice often cannot be transferred to human beings despite the highly physiological 10 system. Experimental Systems V • Special case: Transgene and knock-out animals : Specific elimination of genes (knock-out) or incorporation of genes (transgenes, knock-in) allow a better elucidation of cause-effect relationships. However, classical knock-outs (eliminating a gene in all cells) – is often embryonically lethal – and does not allow conclusions for its function in the adult animal (e.g. mouse). Vice versa, it can happen that there is no phenotype (if the function of the gene is taken over by another gene). Knockins can be quite artificial as well (if the transgene is expressed at higher levels with strong promoters). Modern approaches often aim for „conditional knockouts“ or knock-ins: In most cases the Cre-recombinase / loxP system is used for that purpose: Conditional knock-out: the gene (or a crucial exon) is placed between loxP sites – Cre recombinase (which can be expressed in specific organs or cells by organ specific promoters) cuts out the gene – thus the gene is deleted just in a certain organ or cell type; using inducible Cre, this system allows gene deletion at a defined time point (e.g. when the animal is adult). Conditional knock-in: the gene is placed behind a Stop-cassette, which is flanked by loxP sites. Without Cre activity, the gene is not expressed, with Cre activity (organ specific), the Stop cassette is excised and the gene is expressed 11 5 Conditional transgene mouse models with the Cre / loxP system 1. Cre mouse strain 2. loxP-mouse strain • Cre recombinase cuts out sequences between loxP sites (or inverts sequences between inverted loxP sites) • Cre expression can be rendered cell-type or organspecific using cell-type specific promoters driving Cre expression (spatial control) • Cre expression can be made inducible by using chimeras of Cre with mutated estrogen receptor domains (temporal control: e.g. Cre-ERT2) • Endogenous genes can be flanked by loxP sites (using recombination techniques – e.g. affecting essential exons) > conditional knock-out • Genes can be overexpressed conditionally by inserting an expression construct headed by a loxP-flanked „Stop cassette“, which is cut out by Cre recombinase 12 Examples for conditional mouse models cell-type specific promoter mER Cre mER Cre-ERT2 mER: mutated estrogen receptor (responds to tamoxifen as ligand; without the ligand it keeps the Cre in the cytosol > inactive; upon addition of tamoxifen, the nuclear localization sequences of mER become active leading to translocation of the chimera into the nucleus, where Cre recombinase can exert its function on loxP-flanked DNA sequences). conditional knock-out conditional transgene Gene of interest loxP Stop loxP Gene of interest good promoter (e.g. pCAGGS) mER Tam Cre mER mER Tam Tam Cre mER Tam 13 6 Specific cell ablation or cell labeling in transgenic mice loxP loxP Stop DTR (Diphteria toxin receptor) good promoter (e.g. pCAGGS) cross-breeding with a cell-type specific Cre strain DTR is expressed only in specific cell types injection of diphteria toxin leads to specific killing of these cells loxP loxP Stop good promoter (e.g. pCAGGS) EYFP (enhanced yellow fluorescent protein) > fluorescent labeling of a specific cell type of interest 14 Genome Editing for generating transgene animals • Novel methods to edit genes directly in the genome, e.g. using CRISPR/Cas9 technology or Zn-finger nucleases, or TALENs allow faster and even multiplexing type manipulations (e.g. targeting 5 genes simultaneously) • Genome editing is then usually done in ES-cells, which are subsequently injected into blastocysts. 15 7 Cell Culture Methods • • • • cell culture of immortalized cells cell culture of primary cells (often just a few passages) culture of polarized cells co-culture of different cell types primary fibroblasts (from skin dermis) transformed fibroblasts 16 What you need for cell culture • Incubator (37°C) with CO2supply and humidification • Sterile bench (Laminar Flow: a laminar flow of filtered air keeps the bench sterile) – has to be switched on approx. 10 min before using it, the filter has to be checked from time to time (Particle Measurement) • inverted cell culture microscope (4x, 10x, 20x objectives) • L2-biosafety (e.g. for viruswork): not only incoming air is filtered, but also the air that leaves the laminar flow. 17 8 Culture Media mammalian cells need media with vitamins, amino acids, hormones and growth factors Serum, mostly fetal calf serum (FCS), is the source of growth factors (such as FGF, EGF…) Common composition - DMEM (Dulbecco‘s Modified Essential Medium) - 10% FCS - 2 mM Glutamine (unstable amino acid, has to be added again, if the medium is older than app. 6 weeks) - Penicillin (100 u/ml) - Streptomycin (100 µg/ml) 18 Fetal Calf Serum (FCS) has high concentrations of growth factors (e.g.: EGF, epidermal growth factor; FGF, fibroblast growth factor; IGF, insulin-like growth factor) low amount of antibodies → compatible with cells of other species complement system is inactivated by a heat shock (30 min., 56 °C) disadvantage of FCS: expensive (≈ 100 € per Liter) might contain contaminants such as tetracyclin (important if you use a Tetinducible cell culture system > purchase guaranteed Tet-free FCS) Alternative sources of serum: normal calf serum, horse serum… Special Growth Factors Nerve- Growth Factor (NGF): for neuronal cells Hepatocyte-Growth Factor (HGF): induces cell division of hepatocytes Keratinocyte-Growth Factor (KGF): for culture of skin epithelial cells (keratinocytes) 19 9 Adhesion factors adherent cells versus suspension cells cell culture dishes: usually made hydrophilic (charged groups) – sufficient for most cell types collagen; Gelatine (denatured collagen) sometimes necessary for good attachment (e.g. for primary endothelial cells) Components of extracellular matrix: Fibronectin, Laminin – often better than collagen. special case: „Feeder“-cells (irradiated) – or coculture of cells (e.g. fibroblasts in a collagen matrix with keratinocytes on top) 20 Contaminations in cell culture Bacteria: might be a problem, when they are resistant against the antibiotics that are used (mostly Pen/Strep) > then you have to use other antibiotics (e.g. kanamycin) Mycoplasm: procaryotes of an ancient evolutionary stage, which do not have a normal bacterial cell wall (therefore they are resistant against Penicillin) (can be eliminated with Kanamycin) Yeast: rare (might occur when yeast and mammalian cell culture are not strictly separated) Fungi: quite rare 21 10 Unnoticed mycoplasm contaminations can screw up experimental results > Tests for Mycoplasm-Contamination DNA-staining with DAPI or Hoechst 33258 no Mycoplasm Mycoplasm Alternative: PCR- detection of Mycoplasm-DNA (commercial kits are available) 22 Cell culture of polarized cells apical side tight junction lateral side the cells have to build the tight junctions for building up the polarity. Often they are cultivated on filters, where the two sides are accessible. basal membrane 23 11 Methods to investigate polarized cells Model systems 1. simple cell culture: just one side of the cell is accessible. 2. cell culture on microporous membranes: both sides are experimentally accessible, the establishment of a tight polarized monolayer can be checked be measuring the electrical resistance between the two sides. 3. Organ cultures: e.g. Living Skin Equivalent 4. Organ perfusion: e.g. perfusion of isolated rat liver 24 Culture of polarized cells on membranes Measurement of electrical resistance 25 12 Cultivation of polarized cells on electrode chambers 26 Cultivation of polarized cells on electrode chambers 27 13 Organotypic culture: Example: Skin Equivalent Keratinocytes (cells of the upper layer of skin, the epidermis) are seeded onto a collagen matrix, which contains fibroblasts (dermis cells). As soon as they build a monolayer, they are elevated to the surface of the medium (with their upper side exposed to air) – this induces cell differentiation and the formation of a pseudo-epidermis with several cell layers. 28 Perfusionsdruck (cm H 2(cm O) perfusion pressure Pump Peristaltikpumpe H2O) Liver LEBER bile canula Gallengangskanüle Thermosensor V. porta V. porta Kanüle cavaKanüle canula V.V.cava Computer Fraktionskollektor fraction collector gas humidification Gas-Befeuchtung temperature Temperatur- recording Messung Schreiber Organ Perfusion Polarized cells such as hepatocytes can be maintained in their organ architecture maintaining their polarity. The organ is perfused using glas capillaries linked to the normal blood vessels the supply the organ with blood (and nutrients/oxygen). A buffer (37°C, percolated with O2) containing the nutrients can be perfused through the liver. Marker substances can be applied and their transport from the blood side (basolateral) to the bile side (apical) can be determined. Wasserbad Water Bath Perfusionspuffer Perfusion Buffer 93% O 2 7% CO 2 29 14 Example of an experiment with polarized cells Transmigration of Leukocytes through a Monolayer of polarized endothelial cells Leukocytes are labeled with a fluorescent marker (e.g. CFSE – an ester, which is turned fluorescent just after uptake into cells due to esterases) Cells are seed onto a layer of endothelial cells (e.g. after activating the endothelial cells with inflammatory cytokines – which leads to the synthesis of adhesion molecules on the surface of the endothelial cells). Adhesion of leukocytes leads to transmigration into the lower chamber. The extent of the transmigration can be determined by lysing the cells and quantifying the fluorescence (or by counting the fluorescent cells) 30 Methods for labeling of cellular components • Radioactive labeling: e.g. with radioactive amino acids (35SMethionin…), which are incorporated into newly synthesized proteins. Pulse/chase experiments can give insights into half life, transport. Processing… of proteins. • Internalisation of high molecular weight markers (loading of endosomes and lysosomes). • Labeling of cell surface proteins (e.g. by biotinylation with cellimpermeable reactive biotin compounds). • Addition of cell-permeable labeled substances, which integrate into specific structures (e.g. Golgi-specific fluorescent lipids) • Transfection of cells with expression plasmids • Protein-Transduction (cell-permeable peptides) • Micro-Injection of substances. 31 15 Radioactive Labeling of Biomolecules Amino acids: [35S]Methionine, [3H]Leucin Monosaccharides: [3H]Mannose, [3H]Glucosamine Phosphate: [32P] phosphoric acid Alternative: 33P: lower radioactivity Sulfate: [35S]Sulfuric acid Fatty acids: [3H]Palmitinic acid 32 Radionuclides 33 16 Metabolic labeling with amino acids usually [35S]Methionine labeling advantage: high specific radioactivity disadvantage: relatively rare amino acid > check first, how many methionine are in the protein! (e.g. Ubiquitin: only 1 Methionine, but 9 Leucines) requires Methionine-free culture medium for depletion of endogenous stores by pre-incubation (addition of dialyzed FCS) 34 Metabolic Labeling with Monosaccharides [3H]Mannose or [3H]Glucosamine Activated in the cytosol by coupling to nucleotides → GDP-Mannose, UDP-N-Acetyl-Glucosamine (UDPGlcNAc) requires Glucose-poor culture medium Addition of alternative energy sources (Glutamine, Pyruvate) 35 17 Pulse-Chase-Experiments preincubation without marker (depletion of endogenous stores) Addition of the labeled substance → "Pulse" (5 min 1 h or longer) Stop of the "Pulse" by addition of an excess of unlabeled compound further incubation → "Chase" (min - 48 h) Stop of culture → Analyses 36 Example for a Pulse-Chase-Experiment Fibroblasts of patients, were labeled with 35S-methionine: Pulse: 1 h at 37°C Chase: 7 h at 19°C and 37°C (19°C inhibits transport from trans-Golgi to late endosomes/pre-lysosomes) > Immunoprec. and autoradiography 37 18 Iodination of Proteins (Labeling with 125J) direct: modification of Tyrosine with [125J]: - chemical oxidation: Chloramine-T oxidizes Na125J and leads to iodination of Tyrosine - enzymatic: Lactoperoxidase: oxidizes Na125J in presence of H2O2. indirect: Modification of amino groups (Lysine, NTerminus) with [125J]-labeled Bolton-Hunter-Reagenz (N-succinimidyl 3-(4-hydroxy 5-[125J]iodophenyl)propionate) 38 Biotinylation High affinity binding partner of Avidin and Streptavidin (> easy purification and detection by beads, coated ELISA-Plates…) Streptavidin has 4 binding sites for biotin (complexes can be formed with bivalent biotin-linkers!) > signal amplification is possible (ABC: avidin-biotin-complexes) Most commonly used method to label cell surface proteins Biotinylation of amino groups with BiotinHydroxysuccinimid-Esters or of cysteines (SH-group) by Maleimide-derivates 39 19 Labeling of Endosomes, Lysosomes • By high molecular weight compounds, which are easily detectable, not permeable for the cytoplasmic membrane and taken up by endocytosis Examples: a) unspecific internalization: - FITC-Dextran (Fluorescein-labeled) - Enzymes (Peroxidase…) b) Specifically by receptors Pulse/Chase conditions can be used to labeled specifically early or late endosomes or lysosomes (e.g. using temperature blocks....) 40 Transfections Usually designates the incorporation of DNA into mammalian cells. DNA present in form of plasmids. Transient Transfection: plasmid remains outside of the genome and is slowly lost (degradation, dilution by cell division), exception: episomal replication – e.g. SV40-Plasmids in COS-cells). The transfection efficiency varies – but can reach close to 100% Stable Transfection: integration of foreign DNA into the genome (Efficiency: usually below 0.1%). Isolation of stably transfected clones requires selection genes (for antibiotic resistance, e.g. puromycin, G418…). Plasmids are usually linearized before transfection to increase the possibility of correct integration. 41 20 Example for a mammalian expression plasmid (Replication origins no shown) CMV-Promoter poly-adenylation signal selction gene (bacterial selection with Kanamycin, mammalianselection with G418) reporter gene Multiple Cloning Site target gene SV40-Promoter suited for linearization poly-adenylation signal 42 Selection markers for stable transfections •Aminoglycoside-Phosphotransferase: Resistance against Neomycin (bacteria) und G418 (mammalian cells). Selektion with G418 takes quite long (app 2 weeks). Surviving colonies are isolated and further cultures under selection pressure. •Hygromycin-Phosphotransferase •Puromycin-Acetyltransferase •Dihydrofolat-Reductase (DHFR): Selection with Methotrexat; allows amplifications of the target gene. •GFP-Fusion proteins: fluorescence can be used for selection: example: EGFP-NF-κB expressing CHO-cells 43 21 Chemical Transfection Methods Ca2+ • DNA-Calcium precipitates: at exact pH and Ca2+-concentration: High efficiencies with 293-cells (90% and more), expression levels are usually moderate. Ca2+ Ca2+ • Liposome mediated transfection: kationic lipids which bind the negatively charged DNA, and which are taken up as liposomes Quite high transfection efficiencies but also sometimes toxic effects and potentially artefacts by high expression levels or alteration of cellular membranes. (e.g. Lipofectamine, Fugene...) 44 Buffers • HeBS-Buffer (Hepes buffered saline) 8 g NaCl - 280 mM final concentration 0.2 g Na2HPO4.7H2O (or 0.107 g anhydrous) - 1.5 mM 6.5 g Hepes (Sigma H-7006) (or 5.96 g of free acid) - 50 mM 400 ml A.dest. Adjust the pH to exactly 7.05 (calibrate pH-meter with pH 4.01 and pH 7.00 buffers before). Add A.dest. to 500 ml, filter through 0.2 µm filters and store in aliquots at -20°C (not longer than 6 months). Thawed aliquots shouldn't be frozen again. • CaCl2: 29.4 g CaCl2.2H2O (MW=147) in 100 ml A.dest (final conc.: 2 M) Filter through 0.2 µm filters and store aliquoted at -20°C. • Chloroquine (optional): chloroquine. 2H2O (Sigma C-6628): 12.9 mg/ml in PBS (conc.: 25 mM). Filter through 0.2 µm filters and store at -20°C. Protocoll: Calcium-Transfection Procedure (amounts are given for 6-wells): 1. Seed cells (about 500 000 cells per 6-well = per 10 cm2 ) one day before the transfection (in DMEM/10% FCS) 2. (Optional: 1 h before transfection, exchange the medium for medium containing 25µM chloroquine) 3. Thaw HeBS and CaCl2 at room temperature 4. For each transfection prepare aliquots of 71 µl HeBS 5. Prepare the DNA/ CaCl2-Mix: 4 µg DNA (total) in 62 µl A.dest. + 9 µl CaCl2 6. Add the DNA/ CaCl2-Mix drop-wise to the HeBS aliquots (by screwing the Gilson pipette) and slightly mix after each drop. Incubate for 2 - 3 min at R.T. to form the DNA-precipitate (not longer). 7. Add the DNA-precipitate drop-wise to the cells (by screwing the Gilson pipette and moving it to cover the whole surface of the cell culture; don't swirl the dish). 8. Carefully transfer the dish back to the incubator. Incubate for 24 h (or in the presence of chloroquine: for 10 h) and exchange the medium afterwards. (The transfection is in the presence of FCS!). The efficiency of transfection is in the range of 70-90% for 293 cells. Harvest the cells after 48 h. The protocol is adapted from Neil Perkins who adapted it from Gary Nolan in 1995 (See web site: http://www.stanford.edu/group/nolan/ or CP in Mol.Biol. 9.1 and 9.11.2-3) 45 22 Lipofectamine2000 - Standard conditions Transfection in presence of serum (e.g. 24h – 48h) 46 Chemical Transfection Methodes II • Dendrimeres: charged Polymeres that bind DNA (e.g. Superfect / Qiagen) • complexes with DEAE-Dextran (polycationic Dextran) • poly-ethylene-imine (PEI): 47 23 Physical Transfection Methods • Electroporation: common with suspension cells, electrocuvette requires much DNA Special case: AmaxaNucleofection (now Lonza) • Particle-gun (“Gene Gun”): DNA on gold particles shot by pressure onto cells (e.g. neurons in brain slices). • Micro-Injection of DNA: into single cells (just limited cell number can be targeted). 48 Viral Transfection Methods • Adenoviruses, Retroviruses, Lentiviruses: Viruses developed fancy mechanisms to get into cells – these are applied for gene transfer. Virus-constructs are generated, which contain the target gene but not genes for virus replication (genes essential for generating the viral particles are supplied by “Packaging Cells”). • Adenoviruses: transient Expression • Retroviruses: stable integration but target just proliferating cells • Lentiviruses: stable integration, also transduce quiescent, non-proliferating cells. 49 24 Adenovirus Retrovirus •Episomal gene expression •Long-term, stable gene expression; inheritable •Infects dividing & nondividing cells •Infects dividing cells only •High-level protein expression •Moderate protein expression •Viral titers of up to 1012 pfu/ml •Viral titers of up to 106 cfu/ml (Can be concentrated to 109 cfu/ml) •Accommodates inserts of up to 8 kb •Accommodates inserts of up to 6.5 kb •Elicits immune reactions in vivo •Does not elicit immune reactions in vivo 50 Example of and adenoviral system 51 25 Adeno-associated virus (AAV) • • • • • is a small virus which infects human cells is not causing any obvious disease causes a very mild immune response can infect both dividing and quiescent cells AAV vectors persists mostly in an extrachromosomal state without integrating into the genome of the host cell (the native virus can integrate to some extent into the host genome). • Promising gene therapy vectors (clinical trials have been done for CFTR, hemophilia B, arthritis …) 52 53 26 Protocol: Production of retrovirus by transfection of packaging cells Packaging cell line: Phoenix cells. These cells must not be too confluent since this will lower the production of virus significantly. Day 1 Plate phoenix cells on 15 cm plate (1.5-1.75 x 107 cells) Day 2 ((Transfect phoenix cells using 25 µg DNA and 75 µl Fugene6 + 1.8 ml OptiMEM. Incubate 30 min RT and add to cells.)) OR better use CaPO4!!! Day 3 Carefully remove old medium and add 25 ml of fresh medium. Incubate at 32°C for 24h or 48h (or 72h). Day 4-5 Harvest supernatant after 24-48h. Virus sup can be harvested until the cells start looking unhealthy. Put supernatant in 50 ml tube in ice bucket in the hood. Add 22 ml fresh medium to the packaging cells and put back into incubator. Spin viral sup to pellet any remaining cells. For storage of virus: transfer to cryotube and snap freeze with N2 store at -80°C. Upon freezing virus titer goes down roughly twofold! For use straight away: Filter virus sup through a 0.45-µm cellulose acetate or polysulfonic filter (do NOT use nitrocellulose filter since it binds proteins in the retroviral membrane!). Keep on ice until use. 54 Protocol II: Infection of MEFs with recombinant retrovirus for making stable cell lines Day 3 Split the target cells into 10 cm dishes. Day 4 Add 8 µg/ml polybrene to the filtered viral supernatant. Mix gently by inversion. Replace the media in the target cells with the viral sup + polybrene. Incubate at 37°C for 6h and add equal amount of media containing polybrene. Day 5 Replace the media. Day 6 Replace the media with fresh media containing pyromycin to select for transfectants. The amount of pyromycin used is determined by killing curve experiments. Select colonies of stable transfectants keeping the pyromycin in the media. 55 27 Reporter-Gene Assays Enzymes or other molecules, which are easily detectable, are applied as „reporter molecules“ to detect promoter activities or the activities of signaling pathways. The reporter gene is cloned into an appropriate plasmid (e.g. mammalian expression vector) and transfected into the cells of interest, followed by the biological experiment (e.g. stimulation). Examples for reporter-genes: • • • • • EGFP (Enhanced Green Fluorescent Protein) and variants thereof (e.g. destabilized EGFP) Luciferase CAT (Chloramphenicol-Acetyl-Transferase) ß-Galactosidase (lacZ) SEAP (secreted alkaline phosphatase) etc. 56 Regulated Promoters in Reporter Gene Assays (artificial promoters) • Tandem Repeats of transcription factor binding sites (e.g. 5x NF-kappa B). Often commercially available (e.g. from Stratagene or Clontech) > can be used to determine the activation of a certain transcription factor (or signaling pathway) minimal promoter element Reporter Gene 57 28 Regulated, Natural Promoters in Reporter Gene Assays • Natural promoters usually contain binding sites for several different transcription factors (sometimes several copies of a single TF binding site) > are regulated by several signaling pathways. Example: NFκB IL-8 Promoter IRF-1 GRE AP-1 NF/IL-6 Luciferase p65 / c-Rel > can be used to determine the regulation of a specific promoter of interest Luciferase is usually easier to measure with high sensitivity than the gene of interest (in this case IL-8) 58 Normalization constructs with constitutive promoters …usually used for the normalization control in reporter gene assays (to compensate for differences in transfection efficiency, extraction efficiency or viability of cells in different samples) Usual setting: Pathway-specific reporter construct (e.g. with firefly luciferase) + constitutive normalization construct (e.g. Ubiquitin-Promoter driven βgalactosidase or Renilla-luciferase) > quantification of normalized values (Luciferase / β-galactosidase or Firefly / Renilla-luciferase) Promoters for the normalization vector: • • • • • CMV: from human Cytomegalo-Virus: induces strong constitutive expression (fast, about 1 day after transfection) RSV: from Rous Sarkoma-Virus: weaker, but very constant, constitutive expression, slightly slower (takes 2 d) SV40: Simian Virus 40-Promoter Actin-Promoter: human promoter of a „housekeeping gene“ Ubiquitin-Promoter: human promoter of a „housekeeping gene“ 59 29 Example of an Reporter Gene Assay fold of control vector Luciferase/β-Gal 50 46.36 45 EP-cells 40 DU145 35 26.66 30 25 20 15 10 5 1.94 4.63 0.90 0.49 0 p53 Rb E2F Activities of different signaling pathways or molecules (p53, Rb, E2F) are assessed with reporter constructs containing respective transcription factor binding sites upstream of a luciferase vector. For normalization purposes a constitutively expressed control gene has to be cotransfected (e.g. β-galactosidase downstream of a constitutive promoter e.g. ubiquitin-promoter). Values are calculated as Luciferase/β-Gal. 60 Reporter-Gene Assay Systems for Analyses of Signaling Pathways Transcription factor construct Gene of interest Transcription factor construct ReporterPlasmid In case that the gene of interests activates the specific pathway, which leads to phosphorylation and activation of the used transcriptionfactor construct, the expression of the reporter (e.g. luciferase) is induced. ReporterPlasmid 61 30 Detection of reporter genes: GFP (and variants) • Fluorescence measurement by fluorometry (e.g. in 96-well fluorescence readers) • Microscopy • Flow analysis (cytometry, FACS: Fluorescence activated cell sorting: GFP-containing cells can be separated from other cells and if necessary also further cultivated after purification) GFP fluorescence > 62 Detection of Luciferase (Firefly or Renilla Luciferase) • Luciferin LuciferaseZellextrakt ATP Very sensitive detection in cell extracts by measuring the luminescence generated from luciferase in presence of luciferin and ATP. The substrate Luciferin has to be injected into the sample, and measured immediately (e.g. by integrating for 5 sec) as the emitted luminescence decays quickly. Emitted photons are measured with photomultiplier tubes. PMT Measuring devices: Luminometer (also as 96well devices available) PMT: from http://www.molecularexpressions.com 63 31 Detection of β-Galactosidase (lacZ) • often by photometry (e.g. in ELISA readers) using a yellow substrates, which turns red in presence of Galactosidase (substrate: CPRG, Chlorophenolred-β-DGalactopyranoside). Detection at 595 nm. • Alternative: Chemiluminescence measurement • Alternative: Fluorimetric Detection: Substrate: e.g. 3-carboxyumbelliferyl beta-D-galactopyranoside, CUG is converted to a fluorescent product • Microscopical detection: with X-Gal (5Bromo-4-chloro-3-indolyl-ß-D-galactoside) as substrate: produces a dark blue reaction product 64 β-Galactosidase Assay with CPRG Lyse cells (recommended cell lysis buffer: 0.25M Tris/HCl pH 8.0, 0.25% (v/v) NP40, 2.5 mM EDTA) Pipet about 10 µl of extract into a 96-well plate (appropriate neg. control / blank: 10 µl of mock transfected cells, or non-transfected cells – as there is a slight endogenous b-Gal activity) – leave one well empty for blank (A-1) Add 100 µl substrate solution to the wells (also to the blank-well) Incubate until red color develops (min to hours – depending on b-Gal activity, if you have low activity you can also incubate at 37°C) Optional: Stop with 50 µl of Stop solution (only necessary if you want to time it exactly, e.g. by adding the substrate in a timed way and stopping the reaction in the same way) Measure with ELISA Reader at 570 nm (Filter #3) optimum: 595 nm Lysis Buffer: 0.25M Tris/HCl pH 7.4 (or better 8.0) 0.25% (v/v) NP40 2.5 mM EDTA CPRG-substrate solution: 1 mg/ml (= 1.65 mM) in PBS + 10 mM KCl, + 1 mM MgCl2 alternative substrate buffer: 60 mM Na2HPO4 pH 8.0, 1 mM MgCl2, 10 mM KCl, 50 mM Mercapto-ethanol 65 Stop solution: 0.5M Na2CO3 32 Methods to suppress gene expression (RNA interference) Antisense-Technologies: to suppress gene expression were started many years ago. Early approaches used antisense-oligonucleotides – but the effect was very variable. Alternative approaches used long antisense-strands hybridizing to the mRNA: This usually leads to downregulation of gene expression – but quite often not only for the targeted gene – but also unspecifically for other genes. The reason is that this is „sensed“ by the cells like a long viral dsRNA, leading to virus defense mechanisms: activation of PKR (protein kinase R), phosphorylation of translation factors and general downregulation of protein synthesis. Some years ago, scientists found that small dsRNA in the range of 19-21 nucleotides interferes specifically with target genes, without affecting other genes (small interferent RNA, siRNA) – because they are too small to activate virus defense mechanisms. 66 Principle of RNA-interference Small dsRNA (siRNA) binds to an RNA-induced silencing complex (RISC); consisting of argonaute proteins. Together with the RISC, one RNA strand binds to the target mRNA and leads to specific degradation of this mRNA. The RISC complex is „recycled“ leading to degradation of addition mRNA-targets. 67 33 micro-RNA‘s – the biological mechanism to suppress gene expression miRNAs – can have two effects: 1) mRNA degradation 2) inhibition of translation (this effect often does not require a 100% match with the target mRNA!) Chemically synthesized siRNA Design: traditionally the first AA-Duplett is searched – and the following 19 bases are checked for GC-content (should be 40 – 50%), the sequence should be target gene specific (checked by BLAST) – an appropriate RNA sequence and the reverse complementary RNA are chemically synthesized (and ordered by a company, e.g. Dharmacon, Invitrogen, MWG,…). Company homepages often offer a basic siRNA design: http://www.dharmacon.com/ http://www.mwgbiotech.com/html/s_synthetic_acids/s_rna.shtml The two short RNAs are annealed and transfected (usually using methods that are suited for short oligonucleotides (e.g. Lipofectamine2000 from Invitrogen, XtremeGene from Roche…). 69 34 Vector-coded siRNA > small hairpin RNA (shRNA) •the normal transfection methods, optimized for plasmids can be used •Antibiotics selection genes (e.g. G418, puromycin…) can be included > stable „knock-down“ cell lines can be generated (www.imgenex.com, http://www.oligoengine.com/) 70 small hairpin RNA: hairpin-loop 71 35 Professional Design of siRNA or shRNA • • • • • Design via company website http://www.thermoscientificbio.com/design-center/?redirect=true This delivers a list of several possible sequences (gene specific, checked by BLAST) – with a score based on empirically determined criteria: Nature Biotechnology 22, 326-330, 2004 Check literature for functional siRNA sequences For transduction of primary cells: lentiviral shRNA constructs (also work in non dividing cells) there are also inducible lentiviral constructs available (http://tronolab.epfl.ch/) Many vectors can also be obtained from plasmid repositories: Addgene: http://www.addgene.org Belgian repository: http://bccm.belspo.be/db/lmbp_search_form.php 72 Gene replacement strategy 5‘UTR gene of interest 3‘UTR endogenous mRNA siRNA targeting the endogenous mRNA via the untranslated region Expression plasmid containing: good promoter foreign 3‘UTR mutated gene of interest (SV40 PolyA) the mutated gene replaces the endogenous gene 73 36 Important controls in siRNA experiments • scrambled siRNA as negative control • mutated siRNA with some mismatch as negative control (note: might act as miRNA !) • other siRNAs targeting the same mRNA should have the same effect • if you use shRNA (vector based RNA-interference) use an unrelated shRNA as negative control (e.g. shRNA vector with scrambled siRNA sequence). The empty shRNA vector is not a valid negative control Alternative Methods to influence endogenous protein levels Micro-Injection: This allows injecting antibodies against certain endogenous proteins > interfering with their functions. However, just a limited number of cells (e.g. up to hundreds with automated systems) can be targeted > the following analysis should be a single cellbased assay, such as microscopy. 75 37 Research Methods - Overview • • • • • • • • cell culture systems labelling and transfection of cells analyses of cellular components analyses of molecular interactions fluorescence measurements microscopy flow analysis (FACS) analyses of cellular processes (proliferation, apoptosis..) 76 Analysis of Proteins by SDS-PAGE S-S SO4- reducing agent (DTT) or Mercapto-Ethanol - breaks disulfide bonds 95°C - - - - - - - - SDS – sodium dodecyl sulfate: coats proteins with negative charges - - - - - - 77 38 SDS-Gels For final concentration of gel ( % T): Stack gel Separating gel (10 ml) 30% Acrylamide-bis solution 29:1 (A) (10 ml) 7% 10% 12,5 % 15% 5% 2.33 3.33 4.17 5 1.67 2.5 2.5 2.5 2.5 6x SDS-buffer 4x Separation buffer 1.5 M Tris/HCl pH 8.8 2 M Tris-Cl (pH 6.8) 4x Stacking buffer 0.5 M Tris/HCl pH 6.8 + phenol red aqua dest. 2.5 5 4 3.2 2.4 5.7 0.1 0.1 0.1 0.1 0.1 TEMED 0.015 0.015 0.015 0.015 0.015 APS (10%) 0.03 0.03 0.03 0.03 0.03 SDS (10 %) 2.4 ml SDS 0.96 g Glycerol 4.8 ml DTT 739 mg Bromophenol Blue 4.8 mg 78 Detection techniques Coomassie-Blue staining: robust, moderate sensitivity (limit ≈ 1 µg) Silver staining: elementary silver is deposited at the site of proteins, very sensitive (limit ≈ 10 ng) protein-specific fluorescent dyes: SYPRO-Orange, SYPRO-Ruby, Deep-Purple (compatible with MALDI-TOF, MS) Special stainings: Proteoglycans (Alcian-Blau), glycoproteins (Schiff‘s Reagent) Autoradiographie, Fluorography 79 39 Silver staining 66 kDa 25 kDa separation with 12% Acrylamide: 20 - 80 kDa 80 Molecular weight assessment after SDS-PAGE http://www.meduniwien.ac.at/user/johannes.schmid/SDS-PAGE.xls log[MW] migration distance starting from stacking gel/separation gel interface 81 40 Fluorography, Autoradiography Sensitive detection of radioactively labeled proteins Gel is equilibrated with a radiosensitive fluorophore: e.g. Diphenyloxazole (POP), Sodiumsalicylate Detection by X-ray film or Phoshor-Imager devices 82 Fluorography Phospho-Imager detection (1 day exposure) X-ray film detection (3 months exposure !!) 83 41 Silver Staining of PAGE Gels Solutions Fixing solution: 50 % ethanol, 10 % glacial acetic acid, ad 100 % with aqua dest. Incubating solution (1L): 30 % ethanol, sodiumthiosulfate anhydrous 2g, sodiumacetat anhydrous 34 g, fill up to 1L with aqua dest. Before use add 125 µL of glutaraldehyde/50 mL incubating solution. Silvernitrate solution (1L): AgNO3 1 g, dissolved in 1L aqua dest.. Before use add 10 µL of formaldehyde/50 mL of silver nitrate solution. Developing solution (1L): Na2CO3 anhydrous 25 g, dissolved in 1L aqua dest.. Before use add 10 µL of formaldehyde/50 mL of developing solution. Stop solution (1L): sodium-EDTA 15.78 g dissolved in 1L aqua dest.. After electrophoresis, the polyacrylamide gel is taken out of the casting sandwich and placed in a clean glass beaker filled with fixing solution. All following steps are carried out while gently shaking. The gel has to be incubated with the fixing solution for 30 minutes. After fixation an appropriate amount of incubating solution including glutaraldehyde (the gel has to be at least covered by liquid) is prepared and added to the gel, followed by incubation for 15 minutes, discarding the fixing solution and washing with aqua dest. 3x for 5 minutes and 10 minutes incubation in silvernitrate solution including formaldehyde. The silvernitrate solution is collected (special waste). Developing is carried out by incubating the gel in developing solution including formaldehyde until the desired intensity of protein staining is reached, followed by discarding of developing solution and adding stop solution. The gel should incubate for at least 1 hour in the stop solution. 84 Afterwards the gel can be stored in aqua dest. or dried with vacuum. EMSA‘s (Electrophoretic mobility shift assays) …used to monitor active transcription factors (by binding to short, labeled oligonucleotides comprising the bound DNA sequence) Example: comp.: competitor: non-labeled ds-oligo of the same sequence (usually added in > 10-fold molar excess) – competes with the labeled oligo for binding to the TF > reduces the specific signal mut.comp.: mutated competitor: should not compete for specific binding 42 Defining the composition of TF-complexes using antibodies and supershifts supershift EMSA Alternative: ABCD Assay (Avidin-Biotin Complex with DNA) TF dsOligo Biotin Streptavidin 87 43 Isoelectrical Focussing (IEF) Separation of proteins according to their isolelectric point (pH at which they are not charged) Usage of immobilized pHgradients (Ampholines) native IEF or denaturing IEF (urea) can be done 88 2D-Electrophoresis for analyses of complex protein mixtures (→ Proteomics) combination of IEF (1. dimension) and SDS-PAGE (2. dimension) high separating resolution (> 1000 Spots) Protein pattern databases are available and software for pattern comparison identfication of spots by mass spectrometry or by immunological methods (immunoblotting) 89 44 Principle of 2D-Electrophoresis 90 2D-SDS-PAGE 91 45 2D-DIGE (Difference Gel Electrophoresis) Protein extracts from two different samples are labeled with two different fluorescent dyes (e.g. Cy2 and Cy3) and mixed (usually together with a pooled standard mixture labeled with a 3rd dye, eg. Cy5). 2D-PAGE is performed, followed by scanning of the gel with the 3 wavelengths exciting the 3 dyes > the results are compared by computer analysis Gel chromatography (size-exclusion chrom.) • Separation technique that uses porous bead material in a column to separate macromolecules according to size. • Wide range of molecular weights that can be separated • Much larger molecules can be separated than with SDS-PAGE or native PAGE 93 46 Gel chromatography larger molecules are eluted first 94 Gel chromatography Example 0.017 0.19 SERT Standards 0.015 44 kDa SERT 0.17 0.15 0.11 158 kDa 0.009 0.09 0.07 0.007 0.05 0.005 0.03 670 kDa 0.003 0.01 110 105 95 100 90 85 80 75 70 65 60 55 50 45 40 35 30 25 20 15 5 -0.01 10 0.001 0 Ex436/Em510 0.13 0.011 Trp-fluorescence 0.013 min 95 47 Spin desalting or buffer exchange based on gel chromatography 1. 2. 3. 4. 5. 6. 7. Place spin column in a 1.5 mL microcentrifuge collection tube. Centrifuge at 1500 x g for 1 minute to remove storage solution. Add 300 μL of desired final buffer to the resin bed and centrifuge at 1500 x g for 1 minute; discard flow-through Place the equilibrated spin column into a new collection tube Load your protein sample (130 µl) Centrifuge at 1500 x g for 2 minutes to collect desalted sample. Protein sample is now desalted & buffer exchanged and ready for use 96 Immunoblotting Western Blotting: Electrophoretic protein-transfer from a gel (mostly SDS-PAGE) onto a membrane Membranes: Nitrocellulose, Polyvinyl-difluorid (PVDF) detection via enzyme-coupled antibodies Enzymes: - Horse Radish Peroxidase (HRP) - alkaline phosphatase (AP) 97 48 Immunoblotting II 98 Wet Blotting equipment 99 49 Blotting conditions 100 Semidry blotting (BioRad) • • • • • • • wet Millipore Immobilone-P membrane in MeOH, rinse with water, then 1 min. in Blotting buffer: 25mM Tris Base 150 mM glycine 10% methanol (make a 90% stock without MeOH, which is then added freshly) filter 0.2 mm !!! wet 5 3MM filter papers -> anode then wetted membrane then gel then 5 wetted filter papers wet cathode, close lid quickly without moving back and forth. blot 250 mA for a small Bio-Rad gel (should be 20, increasing to 30 V) for 30 min. 101 50 Immunoblotting: various substrates Chromogenic detection of HRP Chemiluminescence -substrate 102 Western Blot Protocol • • • • • Detection: block 1 hour to O/N in PBS/5% milk at RT wash 3x PBS/0.1%Tween 20 1st AB in PBS/Tween, 1 h wash 3x 5 min PBS/0.1%Tween 20 • add 2nd AB in PBS/Tween (e.g., peroxidase-conjugated donkey anti-rabbit Amersham, at 1:10.000) for 1 hour • wash 3x 5 min ECL-system: • combine equal volumes of soln. A and soln. B (Amersham ECL or Pierce SuperSignal WestPico) • add to filter, incubate 1 min. (Amersham), or 5 min (Pierce) • roll away, cover with saran wrap, expose within 10 min. • (exposure times can vary between seconds and 20 min.) 103 51 ECL reagent: selfmade version Advantages: almost no cost; stable/reproducible, stored in frozen aliquots To 10 mls of 100 mM Tris pH 8.5 (RT), add 50 µl luminol (warm to redissolve) 22 µl of coumaric acid (warm to redissolve) 3 µl of H2O2 (fresh < 6 months) Pour onto blot for 1 minute and process as normal (10 ml enough for 100 cm2) Stock luminol: 250 mM 3-aminopthalhydrazide (Fluka #09253); 266 mgs in 6 mls DMSO; store frozen in 60 µl aliquots. Stock coumaric acid: 90 mM coumaric acid (Sigma C9008): 38 mgs in 2.5 mls DMSO; store frozen in 25 µl aliquots. STRIPPING of membranes: with 2% SDS, 62.5mM TRIS pH6.8, 100mM Beta-mercaptoethanol for 30 min at 50°C 104 Immunoprecipitation Antigen Interaction between antibody and antigen in solution Isolation of immuno-complexes by Protein A- Agarose (Sepharose) – Beads Protein A = IgG-binding protein of Staphylococcus aureus Alternative: Protein G: often used for mouse monoclonal antibodies, which are not well bound by Protein A - or antibody covalently linked to activated Sepharose (CNBr-activated) Analysis by SDS-PAGE 105 52 Example of an Immunoprecipitation Proteins labeled unspecifically with 35S-methionine > specific detection of a protein of interest by immunoprecipitation > fluorography 106 Affinity chromatography • beads can be coupled with antibodies or other affinity ligands, which bind a molecule of interest. • after washing of the beads, the captured target molecules can be eluted e.g. by lower pH. 107 53 Analysis of post-translational modifications proteolytic processing Glycosylation Phosphorylation Ubiquitination …. 108 Analysis of proteolytic processing in vivo: cultivation of cells in presence of selective proteinase inhibitors inhibition of intracellular proteinase (in cytosol, ER, Golgi) is possible just with membrane permeable inhibitors inhibitors that are not membrane permeable act just in the extracellular environment, in endosomes and lysosomes in vitro: Incubation with specific proteinases 109 54 Proteolytic processing (in vivo) Z-FA-CHN2 Z-FA-CHN2 .... Benzyloxycarbonyl-Phenylalanyl-Alanyl110 Diazomethane (inhibits cysteine proteinases) Detection of N-Glycosylation in vivo: Biosynthesis in presence of inhibitors Tunicamycin: inhibits the initiation of N-Glycosylation Processing of N-Glycans blocked by GlycosidaseInhibitors Tunicamycin (µg/ml) 0 0.1 0.5 111 55 Analysis of Glycosylation (in vitro) Endo H .... Endoglucosaminidase H; cleaves just Mannose-rich N-Glycans PNGase ... Peptid:N-Glycosidase F ("N-Glycanase") 112 Analysis of Phosphorylation in vivo: metabolic labelling with [32P]Phosphate (e.g. in presence or absence of an expression or suppression construct for a specific kinase, followed by immunoprecipitation) in vitro: - Incubation with alkaline Phosphatase - Phospho-aminoacid analysis (DC) - Kinase-Assays Immunoblotting/Immunprecipitation (e.g. with anti-phospho- specific Antibody or for instance anti-Phosphotyrosin) 113 56 In vitro Phosphorylation Analysis PNGase ... Peptid:N-Glykosidase F ("N-Glykanase") AlkPhos ... Alkaline Phosphatase 114 in vitro Kinase Assay 1. Immunoprecipitation of the kinase 2. Kinase-Reaction in presence of 3. SDS-PAGE and Fluorography 32P-γ-ATP 32P-γATP + substrate phosphorylated substrate Immunoprecipitated Kinase antibody Agarose-Bead 115 57 Kinase Assay- Protocol Lysis buffer (final conc.): for 20 ml: 20 mM Tris/HCl pH7.5400 µl 1 M150 mM NaCl600 µl 5 M25 mM b-glycerophosphate500 µl 1 M2 mM EDTA80 µl 0.5 M2 mM pyrophosphate400 µl 0.1 M1 mM orthovanadate200 µl 0.1 M1% Triton X-1002 ml 10%1 mM DTT20 µl 1 M1 mM NaF20 µl 1 MA. dest.15.8 ml Protease Inhibitors: added before use (Leupeptin, Pepstatin, Pefa-Block) according to stock Kinase buffer (final conc.): for 20 ml: 20 mM Tris/HCl pH7.5400 µl 1 M20 mM b-glycerophosphate400 µl 1 M100 µM orthovanadate20 µl 0.1 M10 mM MgCl2200 µl 1 M50 mM NaCl200 µl 5 M1 mM DTT20 µl 1 M50 µM ATP50 µl 20 mM1 mM NaF20 µl 1 MA. dest.18.7 ml Lyse cells (in 6 wells) with 500 µl per well of Lysis buffer (+ protease inhibitors): 20 min at 4°C. Clear by centrifugation (14000 rpm, 4°C 15 min Eppendorf centrifuge). Save an aliquot (30 µl) for Western blotting. Immunoprecipitate the kinase (e.g. with 10 µl anti-flag affinity matrix beads, Sigma, for flag-tagged transfected kinase; or with appropriate antibody for endogenous kinase + Protein A-Sepharose or directly coupled to agarose): 2h at 4°C (rotating). Wash the beads: 3x with 1 ml PBS (4°C), 1x with 1 ml Kinase buffer (4°C): pellet the beads by centrifugation (14000 rpm, 4°C, 45sec) and remove the supernatant. Prepare Kinase buffer: add MnCl2 to 10 mM (stock: 1 M) and 32P-g-ATP (5 µCi per sample, usually 1/10 volume, i.e. 1 µl of stock solution for one 10 µl assay) and preincubate at 30°C for 10 min. Add 1 µg substrate: GST-IkB (1 µl) or mutant substrate (as control) to the beads; add 10 µl kinase buffer, mix gently and incubate at 30°C for 30 min (or longer). Stop the reaction by addition of 4x SDS-sample buffer (4 µl) and perform SDS-PAGE with the samples, followed by fixation of the gel (10% methanol, 10% HAc), drying and autoradiography. For detection with PhastGel: use only 5 µl beads, 5 µl kinase buffer, 0.5 µl substrate and 2 µl 4x SDS-sample buffer: Run a 116 12.5% PhastGel with 4 µl per sample Detection of Ubiquitination Potential set-up: • • • • • • transfection of a tagged ubiquitin (e.g. His-tagged) together with the gene of interest (e.g. flag-tagged) immunoprecipitation of the gene of interest optional: resuspend and heat the beads in 1% SDS-buffer, dilute to 0.05% SDS and repeat immunoprecipitation to get rid of potential co-precipitating, interacting proteins. resuspend and heat the beads in SDS-PAGE buffer SDS-PAGE Western Blot for the His-tag 58 Subcellular Fractionation (Separation of subcellular compartments) most often used: density gradient centrifugation coarse separation: differential centrifugation Detection of subcellular compartments by specific markers (enzymes that are nearly exclusively in that compartment) 118 Differential centrifugation Subsequent centrifugation steps with increasing g-force 119 59 Gradient centrifugation: Samples are usually layed on top of a gradient, proteins or compartments migrate through the gradient in „zones“ 120 Density gradient centrifugation Gradients: continuous ↔ discontinuous („steps") self-forming gradients (e.g. Percoll) (density equilibrium centrifugation) samples are either layered on top of the gradient or at the bottom Fractionation after the centrifugation (e.g. by peristatic pump and fraction collector) gradient mixer: B A 0 100 A 100 Magnetic stirrer B 0 volume (or time) 121 60 Substances to generate density gradients Sucrose: low molecular weight (342 Da), osmotically active Ficoll: copolymerisate of Sucrose and Epichlorhydrine; Mr ≈ 400 000 Da Percoll: colloidal silica gel special case for DNA: Cesiumchloride 122 Self-forming gradients (e.g. Percoll..) 123 61 Example for a density gradient centrifugation Separation of lysosomes ( ) and Golgi ( ) in a continuous Percoll gradient (density º) 124 Research Methods - Overview • • • • • • • • cell culture systems labelling and transfection of cells analyses of cellular components analyses of molecular interactions fluorescence measurements microscopy flow analysis (FACS) analyses of cellular processes (proliferation, apoptosis..) 125 62 Methods to investigate macromolecular interactions • • • • • • • Interaction screening with phages (Phage Display) Yeast 1-Hybrid System (protein : DNA) Yeast 2-Hybrid System (protein : protein) Mammalian 2-Hybrid System Gel-Chromatography Co-Immunoprecipitation Fluorescence Resonance Energy Transfer (FRET) – see fluorescence methods 126 Interaction screening with phages (Phage Display) A gene library is expressed on the surface of appropriate phages (e.g. M13), which are incubated with specific target proteins immobilized on plates. Unbound phages are washed off; bound phages are eluted by lowering the pH. 3-4 x Bound phages are amplified and again incubated with plates containing the target proteins – this repeated 3 – 4x to enrich the specifically binding phages. Clones are isolated and sequenced > Sequence of the binding protein 127 63 Yeast 1-Hybrid System For the identification of proteins that bind specifically to a given DNA sequence (e.g. transcription factors; DNA:protein interaction). The DNA sequence of interest (e.g. from a promoter) is usually cloned in repeats (3-5x) in front of an appropriate selection gene (e.g. a histidine synthesis gene) and an appropriate reporter yeast strain (which is not capable of growing in the absence of histine) is stably transformed with this construct. Subsequently, this yeast strain is transformed with a library containing putative binding proteins (often fused to the transactivation domain of the Gal4 transcription factor). Binding of a protein to the DNA sequence results in growth of this yeast clone on selection plates. Gal4-Activation domain Transcription Insert from library TATA HIS > growth on selection plates lacZ DNA-Region with potential protein binding sites (in repeats) 128 Yeast 2-Hybrid System A yeast strain is used, which does not contain a functional Gal4 transcription factor – but reporter and selection genes, which are downstream of Gal4-dependent promoters (Histidine- and Adenine-synthesis genes, lacZ for β-Galactosidase expression, which can be used for staining). This strain is transformed with putative interaction partners: 1. fusion protein of the Gal4-DNA-binding domain and Protein X („bait“) 2. fusion protein of the Gal4transactivation domain and Protein Y (or a library insert; = „prey“) Co-Transformation or combination by yeast mating > In case of an interaction between protein X and Y auftritt, a functional transcription factor is build, which binds to Gal4 promoters – and the cells can grow on selection plates (without His or Ade). 129 64 Preparation of a Yeast 2-Hybrid Screen 1. Cloning of the gene of interest into the bait vector“ (in frame with the Gal4-DNAbinding domain): selection in bacteria (e.g. via Kanamycin), selection in yeast (e.g. via Trp-synthesis gene)) 2. Test for Auto-Activation with „empty Gal4AD-vector): Tests whether the gene of interest interacts with the Gal4 activation domain (without the need of protein Y): If it does so, the bait cannot be used in the yeast 2-hybrid system. The Gal4 AD vector contains a second selection gene for yeast (e.g. leucine synthesis gene). Co-transformants of the two vectors grow in the absence of Trp and Leu; but they should not grow in the absence of the amino acid that is synthesized just when an interaction occurs (e.g. in the absence of histidine or 130 adenine). Combination of the two putative interaction partners Reporter strain Y187 pretransformed with a library (in Gal4AD-Vector), Mating Type: α Reporter strain (e.g. AH109) Mating Type: a bait X Gal4AD Gal4BD Mating: Incubation of the two haploid strains for 24 h at 30°C, 40 rpm > formation of diploid clones with both vectors clones, which contain interaction partners grow on selection plates and express lacZ Instead of mating the two vectors can be combined by classical transformation X His, Ade, lacZ PCR from single colonies (with primers specific for the library vector) purification of PCR-Products sequencing of the putative interaction partners 131 65 Example of a yeast two-hybrid result A) single colonies on selection plates (SD-Leu-Trp-Ade) B) Streaking out the colonies from the first selection plates to secondary selection plates (e.g. with higher selection pressure and stringency: SD-Leu-Trp-Ade-His) 132 Verification of a yeast 2-hybrid result 1. Analysis of the sequence and comparison with database: check whether the ORF is OK (in frame with the Gal4AD) 2. Isolation of Plasmid-DNA from the yeast colony 3. Re-transformation in E.coli (to separate bait and prey – using different antibiotics resistance) and preparation of the plasmid containing the library insert 4. „False Positive Test“ in yeast: Transformation of the Gal4AD-plasmid containing the identified „prey“ with the empty Gal4-binding domain vector: this shouzld not lead to growth on selection plates of interaction (if there is growth, then the library insert interacts with the Gal4BD and not the bait protein) 5. β-Galactosidase-assays (also quantitative, to compare interaction partners) 6. Verification in the correct cells (human cells), e.g. by co-immunoprecipitation 133 66 Example of a “False-Positive Test” 1,7 1,6 rel. activity 1,5 Example of lacZ quantification 1,4 1,3 1,2 1,1 1 neg. control IKK2/GMRa IKK2/GMRb 134 LiAc Yeast Transformation • • • • • • • • Solutions: Synthetic drop out solution 10 x in AD: L-isoleucine 300 mg/L, L-valine 1.5 g/L, L-adenine hemisulfate salt 200 mg/L, L-arginine HCl 200 mg/L, L-histidin HCl monohydrate 200 mg/L, L-leucine 1 g/L, L-lysine HCl 300 mg/L, Lmethionine 200 mg/L, L-phenylalanine 500 mg/L, L-threonine 2 g/L, L-tryptophan 200, L-tyrosine 300 mg/L, Luracil 200 mg/L SD -Trp medium (synthetic dropout medium): synthetic minimal medium lacking tryptophan: yeast nitrogen base without amino acids 6.7 g/L, 2 % dextrose (glucose) (sterile dextrose solution is added after autoclaving to avoid maillard reactions), pH adjusted to 5.8, for plates : agar 1.5 g/L YPD (yeast peptone dextrose) broth, yeast complete medium: yeast extract 10 g/L, peptone 20 g/L, 2 % dextrose (glucose), pH adjusted to 5.8 DNA + water + carrier-DNA or RNA Aqua dest. sterile 1M LiAc LiAc 100 mM sterile Polyethylenglycol LiAc 1 M sterile Bacterial RNA, used as carrier yeast pellet Poly-ethyleneglycol PEG 50 % (w/v) sterile filtered 10 mL of SD -Trp medium are inoculated with the appropriate yeast strain and incubated at 30°C while shaking at 200 rpm o/n. On the next day OD at 600 nm is measured and the yeast culture is diluted with YPD to OD600 0.1. A total volume of 50 mL diluted yeast culture is used for further incubation. Every hour OD600 is measured until OD600 0.4 is reached (3 - 5 hours). Then the cell number is calculated with a Thoma chamber. 2x107 cells/mL are sufficient for 10 transformations. The yeast is then harvested by centrifugation at 3000 rpm for 5 minutes, the supernatant is carefully removed and collected for autoclaving. The pellet is resuspended in 25 mL sterile AD and again centrifuged at 3000 rpm for 5 minutes. After removing of the supernatant the pellet is resuspended in 1 mL LiAc 100 mM. Excess of LiAc is removed by spinning the tubes for 15 seconds at full speed in a tabletop centrifuge and carefully removing the supernatant. The yeast pellet is brought to a final volume of 500 µL with LiAc 100 mM. Aliquots of 50 µL are prepared. One 50 µL aliquot of this yeast suspension is used for one transformation. 50 µL aliquots are again briefly centrifuged to pellet the cells, the supernatant is removed and on top of the yeast pellet, layers of the following transformation solutions are pipetted in following order: 240 µL 50 % PEG, 36 µL LiAc 1 M, 3.3 µL of bacterial RNA (31 µg/µL), 70.7 µL sterile AD, 1 µg plasmid DNA (1µg/µL). The tube is then thoroughly mixed by vortexing for 1 minute until the yeast pellet is completely dissolved and placed for 30 minutes in a 30°C water bath. The tube is then transferred to a 42°C water bath for 25 minutes in order to perform the heatshock. The transformation mix is then briefly centrifuged for 15 seconds at 4 000 x g (7 000 rpm) in a table top centrifuge, the supernatant is discarded and the pellet is resuspended 135in 1 mL sterile AD. 50 µL of this transformed yeast suspension are plated on SD – Leu, - Trp, - Ade plates and incubated at 30°C for some days. 67 Mammalian 2-Hybrid System Posttranslational modifications such as phosphorylations, which might be essential for interactions are often not carried out in yeast. In this case a similar assay can be set up in mammalian cells (e.g. providing the kinase) Limitations: 1. not suited for screening purposes 2. Proteins are in the nucleus and thus eventually not at their normal localization > FRET-Microscopy: as alternative to visualize protein-protein-interaktion in their physiological context 136 Example for a mammalian 2-Hybrid Test 137 68 Biochemical Verification of Protein-Interactions by Co-Immunoprecipitation (CoIP) 1. The 2 proteins of interest are transfected into mammalian cells (usually containing to different tags, e.g. HA- and flag). 1 – 2 d after transfection, the cells are lysed and one protein is immunoprecipitated using antibody-beads against tag1 (e.g. flag). The beads are washed extensively with buffer (isotonic or hypertonic, not hypotonic) and finally heated with SDS-buffer to release bound proteins. SDSPAGE and Western blotting is performed – using antibodies against tag1 and against tag2. If protein with tag2 co-precipitated with protein containing tag1, then there is interaction. 2. Co-immunoprecipitation of endogenous proteins (without transfection) – using the same principle and antibodies against the endogenous proteins 1. Protein X-myx Protein Y-HA 3. 2. HRP Antikörper bead 138 Example for a Co-Immunoprecipitation 1. Co-IP with overexpressed proteins (after transfection); control: transfection with just one protein 2. Co-IP with endogenous proteins control: IP with unrelated antibody 139 69 The salt concentration has an influence on the stringency of the co-immunoprecipitation Co-Immunoprecipitation of TRAF1 and TRAF2 at 500 mM NaCl Co-Immunoprecipitation of TRAF1 und IKK2 works at 250 mM NaCl, but not at 500 mM > TRAF1/TRAF2 interaction is stronger than TRAF1/IKK2 interaction 140 Co-Immunoprecipitation for the Detection of Protein Interactions 1. Transfection of cells with tagged proteins (one 6-well of CHO or HeLa cells is sufficient for one sample). 2. Preparation of extracts: 2.1. 1 d after transfection: wash cells with PBS 2.2. Lysis with 500 µl/well Lysis-Buffer + Protease Inhibitors: 15 min at 4°C. Buffer: 0.5% NP40, 50 mM Tris/HCl pH 7.5, 1 mM EDTA, 150 mM NaCl. Protease Inhibitors: 10 µg/ml Aprotinin, 20 µg/ml Phosphoramidon, 40 µg/ml Pefabloc, 1 µg/ml Leupeptin, 1 µg/ml Pepstatin (from 1000x stock solutions, Boehringer Protease Inhibitor set).The lysis is suited for cytosolic proteins and membrane proteins. Nuclei remain intact (you can leave the nuclei on the plate when you take off the supernatant). 2.3. Spin the extracts for 15 – 30 min at 14 krpm, 4°C (HeLas: 15 min, CHO: 30 min) 2.4. Keep the supernatant and adjust the NaCl-concentration (150 mM – 1000 mM depending on the strength of interaction; start in the range of 150 – 250 mM, increase the concentration if you want to increase the stringency) 3. Co-Immunoprecipitation 3.1. Take 400 µl of extract for IP (keep about 30 µl extract for direct western analysis). Use flat-top tubes (the visibility of the pellet is better in these tubes) Add 400 µl Lysis-Buffer/250 mM NaCl (without NP40 > final concentration: 0.25%). Add beads (15 µl anti-flag-M2-Agarose, Sigma A-1205; alternatives: other antibodies directly coupled to CNBr-activated Sepharose; Protein A- or Protein G-Agarose: the later will give more unspecific binding). Rotate extracts + beads for 2 h at 4°C. 3.2. Spin for 30 sec at 14 krpm 4°C. Take off the supernatant, add 1 ml of lysis buffer/250 mM NaCl/without NP40 and invert tubes several times (do not vortex). Repeat this washing step. 3.3. Suspend the beads in 1 ml cold PBS and transfer the suspension to a new tube. Spin 30 sec at 14 krpm, 4°C, take off the supernatant and repeat this washing step. Final centrifugation: 1 min at 14 krpm, 4°C. Remove the supernatant and suspend the beads in SDS-PAGE buffer (30 µl). Incubate for 5 min at 95°C and pellet the beads for 2 min at 14 krpm. 4. SDS-PAGE 5. Western Blot: if possible use HRP-conjugated primary antibodies (anti-HA-HRP from Boehringer, anti-mycHRP from Invitrogen). This gives much lower background of unspecific bands (Ig light chain …). 141 70 Far Western Blotting … the membrane is probed with a protein, which can bind the protein of interest. While western blotting detects certain proteins using antibodies, farwestern blotting detects protein:protein interactions. Research Methods - Overview • • • • • • • • cell culture systems labelling and transfection of cells analyses of cellular components analyses of molecular interactions fluorescence measurements microscopy flow analysis (FACS) analyses of cellular processes (proliferation, apoptosis..) 143 71 Principle of Fluorescence 1. electrons of a fluorophore are excited by absorption of an appropriate photon (hν −Ex) and their energy state is raised to S1´ Jablonski-Graph 2. the excitated state S1´exists for about 1 – 10 nsec. Energy is lost by several reactions (interaction…) leading to the excited state S1. 3. Electrons fall back from S1 to S0 – the energy difference is released by emission of a photon (which has lower energy than the excitation photon – and thus a longer wavelength) according to λ = c/ν 144 Principles of Fluorescence double bonds = flexible (delocalized) p-electron system from: http://www.invitrogen.com/site/us/en/home/support/Tutorials.html 72 delocalized p-electron systems (alternating double bonds) can easily absorb photons and thereby be raised to higher energy levels delocalized p-electron systems (alternating double bonds) can easily absorb photons and thereby be raised to higher energy levels 73 energy loss due to movements, rotations etc... ... sudden fall from an excited energy level to the ground state 74 The absorbance of light (photons) depends on the colour (the wavelength) number of absorbed photons excitation spectrum 75 The emitted light (photons) exhibits a certain wavelength spectrum (colour) – depending on the nature of the fluorophore number and colour of emitted photons emission spectrum 153 76 Excitation and Emission Spectra Stoke‘s Shift Infos: http://www.probes.com/servlets/spectra/ Java-Applet from BD: http://www.bdbiosciences.com/spectra/ 154 Java-Applet from BD: http://www.bdbiosciences.com/spectra/ 155 77 Characteristics of fluorescent dyes Excitation Maximum: wavelength of maximal photon absorbance (λ in nm) Emission Maximum: wavelength of maximal photon emission (fluorescence, λ in nm) Molar Extinction coefficient: gives the absorbance of excitation photons at the excitation maximum λ (in cm1M-1) Quantum-Yield: number of emitted photons per number of absorbed photons. Brightness = molar extinction coeff. x quantum yield 156 Parameters of some important fluorescent dyes dye Ex Em DAPI 359 461 FITC 494 520 TMRho 550 573 TexasRed 595 615 DAPI... 4’,6-Diamidino-2-Phenylindol FITC... Fluorescein Isothiocyanat TMRho...Tetramethylrhodamine (TRITC: Tetramethylrhodamine Isothiocyanate) 157 78 Alexa-Fluorophores from Molecular Probes/Invitrogen (www.invitrogen.com) 158 GFP (Green Fluorescent Protein) and its variants • Structure: barrel like with the chromophore in the middle • MW: appox. 29 kDa • original protein from jellyfish (Aequorea victoria), exists in bacterial and mammalian codon optimized versions. • Point mutations were incorporated improving the fluorescence (enhanced GFP: EGFP) and also leading to other spectral variants (colours; ECFP, EYFP…) • Fluorescent in living cells – can be expressed as fusion protein with the protein of interest (is usually not altering the function of the target protein) 159 79 Fluorescence Properties of some GFP-Variants Variant Excitation (nm) Emission (nm) EBFP (Blue) 380 440 ECFP (Cyan) 433 475 EGFP (Green) 488 507 EYFP (Yellow) 513 527 DsRed 558 583 160 Other fluorescent proteins fluor. protein Ex-Peak nm Em-Peak nm quantum yield comment EBFP 380 440 0.18 ECFP 433 (453) 475 (501) 0.4 Clontech Clontech EGFP 488 507 0.6 Clontech 397 (475) 509 0.77 Aequorea victoria EYFP 513 527 0.61 Clontech Citrine 516 529 0.76 Griesbeck et al. 2001 DsRed 558 583 0.29 Clontech, tetramer 0.55 tetramer, wildtype GFP DsRed2 563 582 588 618 0.02 Clontech, dimer PA-GFP (Patterson 2002) 400 before act. 504 (397) after 515 before act. 517 after act. 0.13 0.79 photoactivatable GFP, T203H mutant of mammalian codon-optimized wildtype GFP PS-CFP 400 before act. 490 after act. 468 before act. 511 after act. 0.2 0.23 photoswitchable CFP, turns from cyan to green after intense illum. at 405 nm mOrange 548 562 0.69 Shaner et al., 2004 596 0.29 Shaner et al., 2004 610 0.22 Shaner et al., 2004 581 0.69 Shaner et al., 2004, dimeric HcRed1 mStrawberry mCherry dTomato 574 587 554 161 80 Photoconvertible fluorescent proteins mOrange conversion to far-red (2x bleaching with 100% 488 nm in between) 120 100 rel fluor 80 mOrange 60 converted far red 40 control cell 20 0 0 20 40 60 80 100 120 sec mOrange Far-Red Photo-switchable fluor. protein Dronpa 120 start bleaching at 488 nm % of initial fluor. 100 80 60 start reactivation at 350 nm 40 20 0 -20 0 20 40 60 80 100 120 140 160 180 200 sec 81 Fluorimetric Analysis Methods Fluorescence measurements are usually more sensitive than photometric measurements. Scanning fluorometers have usually 2 monochromators for adjusting excitation and emission wavelengths. Most instruments also allow to adjust the bandwidth of excitation and emission (between 1–20 nm) The emitted fluorescence is measured by a photomultiplier tube (PMT) detector. The sensitivity of that can be adjusted by changing the voltage (e.g. 400 - 700 V). Emission Monochromators Detector 164 Parameters of fluorometry • • • • • • Excitation wavelength in nm bandwidth of the excitation light (1 – 20 nm, „slit width“) Emission wavelength in nm bandwidth of the emission sensitivity of the detector („gain“, voltage of the PMT) Integration time of the measurement (slow – fast, in sec.: influences the „noise“) 165 82 Wavelength Scans The exact emission and excitation peaks might differ slightly between different fluorometers. Emission spectrum (ECFP) 2 For checking the parameters: - run an excitation wavelength scan at the literature value of the peak emission - run an emission scan at the determined excitation peak - repeat the excitation scan at the determined emission peak rel. fluor. 1.5 1 0.5 0 460 480 500 520 540 nm 560 580 For adjusting these parameters you have to consider the fluorescence properties (e.g. the Stoke‘s shift) – to prevent that excitation light is detected 166 Tricks for optimizing fluorescence measurements based on the spectra excitation window possible emission window Fluor. real emission curve 1 theor. excitation curve spill-over of the excitation light theor. emission curve detected fluorescence nm 83 Tricks for optimizing fluorescence measurements based on the spectra Fluor. excitation window more narrow emission window theor. excitation curve emission curve 2 detected fluorescence nm Tricks for optimizing fluorescence measurements based on the spectra left shifted broader excitation window Fluor. real emission curve 1 real emission curve 3 detected fluorescence nm 84 Time Scans Can be applied to determine the time course of fluorescence changes (e.g. to determine enzyme reaction kinetics – or for instance in chromatography to measure the kinetics of elution and thus the molecular weight of a fluorescent compound, such as a GFP-fusion protein) 0.017 0.19 SERT Standards 0.015 44 kDa SERT 0.17 0.15 Ex436/Em510 0.13 0.011 0.11 158 kDa 0.009 0.09 0.07 0.007 Trp-fluorescence 0.013 0.05 0.005 0.03 670 kDa 0.003 0.01 110 105 95 100 90 85 80 75 70 65 60 55 50 45 40 35 30 25 20 15 5 10 -0.01 0 0.001 170 min Fast Kinetic-Analysis (Stopped-Flow Fluorometry) Two reaction partners are injected into a mixing chamber, where they are mixed within approx. 1 msec by stopping the flow. If the reaction between the two compounds changes the fluorescence, this change can be recorded with a resolution in the microsecond range. Light-Absorbance (Stopped Flow Photometry) A Fluorescence (Stopped Flow Fluorometry) B Excitation light 171 85 Example for a stopped-flow fluorometry 0.008 0.007 0.006 0.005 0.004 0.003 0.002 0.001 0.000 -0.001 -0.002 Linear correlation between the initial reaction kinetics range and the concentration of the reaction partners Tet-DNA mutant Tet-DNA 0 0.02 0.04 seconds 0.06 rate (1/sec) rel. fluorescence Tangent of the initial fluorescence change (reaction kinetics rate in 1/sec) kon koff y = 734,21x + 244,09 R2 = 0,9508 900 800 700 600 500 400 300 200 100 0 rate (15°C) rate (37°C) kon koff 0 0,2 Kaff = 1/Kdiss = kon / koff 0,4 0,6 0,8 DNA (µM) 172 Quantitative Fluorometry Fluorescence measurements can be used to quantify a great variety of different substances. Usually a standard curve is measured with the optimized measurement parameters (e.g. after defining them by wavelength scans: excitation and emission wavelengths and corresponding bandwidths; PMT voltage and integration time). Some examples for quantitative fluorometry 1. Enzyme-Measurements (e.g. β-Galactosidase) 2. DNA-Measurements (Hoechst 33258, SYBR Green): fluorescent dyes, which intercalate into the DNA and are fluorescent dependent on the amount of DNA 3. Protein-Measurements (inherent fluorescence due to aromatic amino acids such as Tryptophane (dye: SYBR Orange, …) 173 86 Example for quantitative fluorimetric measurement Fluorescence (Em 376/ Ex 276) 7 Standard curve 6 Probe 5 4 3 2 y = 0.8738x + 0.1417 2 R = 0.9959 1 0 0 1 2 3 4 5 6 7 8 µg/ml 174 Fluorescence as measurement value in special analysis techniques: Real-Time PCR Conventional PCR: Comparison of a gene of interest with a housekeeping gene using an endpoint determination Sample 1 In real: samples contain different amounts of cDNA, but this difference is not detected, when they reach the same plateau at the end of the reaction > this can be revealed by measuring the reaction product (by fluorescence) after each cycle 2 1 endpoint 2 175 87 PCR Principle 176 177 88 178 Scheme of a qPCR machine LightCyclerTM, Roche heating Ventilator capillary with the PCR-mix dichroic mirrors threshold Ct light source detectors 179 89 Realtime PCR machines Applied Biosystems Roche Light Cycler StepOne Plus capillaries (app. 1 €/sample) 96-well plates 180 Real-Time PCR with SYBR Green as DNAfluorescence dye SYBR Green intercalates in the amplified dsDNA (PCR-product)leading to an increase in fluorescence with increasing cycle number. After many cycles the fluorescence also increases in the water control – due to the formation of primer aggregates 181 90 Melting point analysis of the PCR-product Specific and unspecific PCR products can be distinguished by their different melting temperature (as determined after the PCR by slow heating and the decrease of the fluorescence at the melting point). This can also be applied to detect mutations. measurement of the fluor. after each cycle at a temperature above the melting temperature of the unspecific PCR product allows quantifying just the specific product 182 Real-Time PCR with FRET-Hybridisation Probes Within the sequence flanked by Primer 1 and Primer 2 (amplification primers), two additional oligonucleotides (Hybridisation Probes 1 and 2) are situated, which contain two different fluorophores at the 3‘ and 5‘ ends. When these oligos bind to the PCR product, the fluorophores come into close proximity and one fluorophore can transfer part of its fluorescence energy to the other one (fluorescence resonance energy transfer, FRET), which then starts to shine. In this case primer aggregates do not generate a fluorescence signal. 183 91 Real-Time PCR with TaqMan probes A TaqMan-probe contains FRET-Donor and Acceptor (Quencher)-Fluorophore within the same oligonucleotide. At the annealing temperature of the oligo, the probe binds to the PCR product. The exonuclease activity of the Taq-Polymerase cleaves the probe and leads to increase of the Donor-fluorescence due to de-quenching. 184 Realtime PCR – Quantification of gene up/down-regulation PCR efficiency CP 1. Determine the PCR efficiency of your gene of interest and that of your housekeeping (reference) gene using serial dilutions (e.g. of plasmids or cDNA preparations): E = ideally 2 (duplication at each cycle) but realistically lower (e.g. 1.8) 35 33 31 29 27 25 23 21 19 17 15 y = -1.67ln(x) + 25.767 R² = 0.9972 crossing point CP Log. (crossing point CP) 0.1 2. Calculate up- or downregulation of your specific gene of interest using the differences in the crossing point (CP) values with the equation: ratio = ( Et arg et ) ( Eref ) 1 10 100 ng cDNA input ∆∆Ct-method (Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 2001, 29(9):e45 ∆CPt arg et ( control − sample ) ∆CPref ( control − sample ) (Excel template on my website) 92 100000 Calculating the PCR efficiency from the shape of the curve log (Fluor) 10000 1000 100 - Without using a dilution curve 10 0 10 20 30 40 50 cycle - Can be calculated for each sample separately 80000 70000 Neurosci Lett. 2003 Mar 13;339(1):62-6. 60000 Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Fluor 50000 40000 Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 30000 20000 10000 0 0 -10000 10 20 30 40 50 (Excel template on my website) 186 cycle LinRegPCR Software http://www.hartfaalcentrum.nl/index.php?main=files&sub=LinRegPCR 187 93 188 Research Methods - Overview • • • • • • • • cell culture systems labelling and transfection of cells analyses of cellular components analyses of molecular interactions fluorescence measurements microscopy flow analysis (FACS) analyses of cellular processes (proliferation, apoptosis..) 189 94 Microscopy: Human vision and the concept of magnification image formation in the human eye 2-step magnification principle of a microscope with 2 lenses: objective and eye piece (occular) 190 Basics of optical resolution I Fine structures induce a diffraction of light (light of zero-order, 1st order ...). Light diffraction on a small iris is more or less equal to diffraction on small cellular structures: sinθ(1) = 1.22(λ/d) θ ... angle to the first light minimum λ... wavelength d ... diameter of the iris for very small angles θ: θ(1) ≅ 1.22(λ/d) objects that are closer than θ(1) cannot be resolved as separate objects 191 95 Basics of optical resolution II The more orders of light are resolved the better is the resolution. The optical resolution that can be achieved is defined by the so called numerical Aperture (N.A.) of the objective. N.A. = i sin q i ... Refraction index of the medium (e.g. 1.0 for air, up to 1.56 for oil) q... half of the objective opening angle (Aperture) 192 „Airy“ disks: optical basic structures 193 96 Deceleration (phase shift) of light by passing through an object http://www.microscopyu.com/tutorials/java/phasecontrast/phasespecimens/index.html 194 Köhler Illumination … was established to guarantee optimal illumination of objects. This illumination is usually also a prerequisite for different contrast methods (phase contrast, differential interference contrast) to work, as the necessary components are optimized for Köhler illumination. In order to get a Köhler illumination, you have to focus the object first, then you close the field iris, so that just the middle part of the view field is illuminated (if necessary you have to center the light path) and then the vertical position of the condensor is adjusted so that the borders of the field iris appear sharp and focussed. 195 97 Correct and wrong illumination field iris at wrong vertical position of the condensor light path not centered Correct Koehler illumination 196 Specifications of objectives • • • • • • Magnification (10x, 20x, 40x…): total magnification is given by objective magnification and occular magnification (or magnification of the lens in front of the CCD camera) Immersion medium: air, oil – or water High magnification (40x – 100x) with oil or water additional features (e.g. suitability for fluorescence due to low autofluorescence of the glass: Neofluar..) contrast features: e.g. phase contrast (e.g. Ph1) correction of lenses for chromatic abberations (e.g. Apochromat) correction of lenses for planarity of focus over the view field (Plan) 197 98 Contrast enhancement in transmitted light microscopy 1. Staining of structures (e.g. nucleus: blue with hematoxylin, antigen: brown with immunohistochemistry) 2. Phase contrast: Making use of the phase of light when it passes an object 3. Differential-Interference contrast (Normarski): Making use of light polarization and its change through objects to generate a contrast 198 Phase contrast Unstained objects such as cells slow down the light (the phase of the passing light) by ¼ λ. Phase contrast rings in the objective can accelerate the light, which does not pass through cells by ¼ λ, the resulting difference of ½ λ causes an interference, which leads to contrast enhancement. ¼λ ½λ http://www.microscopyu.com/tutorials/java/phasecontrast/opticaltrain/index.html 199 99 Phase contrast II The phase contrast rings of the objektive and the condensor have to match each other in diameter and have to be concentric. In addition the distance between them has to be correct (which is the case at the Köhler illumination) – this is especially important for higher magnification objectives. It is stated on the objektive which phase contrast ring has to put in at the condensor (e.g. Ph1, Ph2...). 200 phase contrast III wrong phase contrast ring correct Koehler illumination and phase contrast phase contrast rings not centered 201 http://www.microscopyu.com/tutorials/java/phasecontrast/microscopealignment/index.html 100 Illumination scheme of an inverted microscope field iris condensor with phase contrast rings Knob to adjust the vertical position of the condensor screws for centering the light path 202 Differential-Interference-Contrast (DIC) (Normaski Contrast) Before reaching the condensor, the light is polarized and passes a double prism (Wollaston Prism), where it is split into two beams with different directions and perpendicular waves. These beams pass the sample, where they are altered in intensity and phase etc. The beams are focussed by the objective. In the focal plane there is a second double prism, which combines the beams again. After that, the beams are depolarized again. Thereby the beams that have been altered differentially in the sample can interfere with each other – and this interference results in changes of the intensity and the colour. The outcome is a preudo-threedimensional image. 203 101 Comparison Phase Contrast – Differential Interference HeLa cells – same view field http://www.microscopyu.com/tutorials/java/phasedicmorph/index.html 204 Fluorescence Microscopy Example: TripleFluorescence-labeled endothelial cells: Red: Actin-Filaments labeled with Phalloidin Green: Membranes (DiO-C6) Blue: Nuclei (DAPI-staining of DNA) 205 102 Basics of fluorescence microscopy Fluorescent samples are excited with light of an appropriate wavelength (through the objective), the emitted fluorescence is collected again by the objective and is guided to a dichroic mirror, which separates the excitation light from the emitted fluorescence; the latter passes an emission filter and is detected (by eye or by appropriate detectors such as cameras) Interactive Zeiss-Tutorials: http://zeiss-campus.magnet.fsu.edu/tutorials/ 206 Scheme of a fluorescence microscope 207 103 Light sources for fluorescence excitation 1. Conventional light sources: - mercury lamps: - Xenon-lamps: 208 LED Light Sources (light-emitting diodes: semiconductor devices) from: http://zeiss-campus.magnet.fsu.edu/ 104 Metal Halide Lamps from: http://zeiss-campus.magnet.fsu.edu/ Laser light sources 2. Laser (Light Amplification by Stimulated Emission of Radiation): Give just discrete wavelengths (lines) – thus the choice of excitation light is limited and depends on the laser type. Ar-laser: main lines at 488 nm and 514 nm, (and 458) He/Neon: 543 nm, 633 nm UV-Laser: 405 nm Violet laser diodes: 405 – 420 nm Advantages of laser light: -high quality (parallel light beams) -good for scanning -high intensity 211 105 Fluorescence Filter Cubes The filter cube consists of: 1. Excitation filter: just the correct excitation light (wavelength) passes the filter 2. Dichroic mirror: is reflective for the excitation light but transmittent for the emission light (the emitted fluorescence) – separates excitation from fluorescence light sample 3. Emission filter: filters the emitted light so that just the correct wavelength (e.g. in double fluorescence) reaches the detector 212 Characteristics of fluorescence filter sets Excitation filter (Band Pass) Emission filter (Band Pass) dichroic mirror excitation filter emission filter 213 106 Example of a bandpass filter + nomenclature 214 Dualband filter sets: Simultaneous observation of two different fluorophores (e.g. EGFP/DsRed) Excitation dichroic mirror Emission 215 107 Monochromators as light source A conventional light source (e.g. a Xenon lamp) is split by a monochromator (e.g. a diffraction grid) to the spectral colours – to produce light of a freely definable wavelength (320 – 700 nm). This can be used instead of a fixed excitation filter. One advantage is that this technology allows switching between different excitation wavelengths within few milliseconds. This can be important for excitation ratio imaging (e.g. Fura-2 Calcium imaging etc.) UV (320 nm) electronically adjustable grid Polychrome V from TILL Photonics Red (700 nm) 216 Detection of the emitted fluorescence - Visually via the occular of the microscope - by a CCD camera (usually cooled to reduce the electronic noise). The photons of the fluorescence hit a light sensitive chip (e.g. out of 1300 x 1030 pixels), where electrons are released dependent on the intensity of the fluorescence. Each chip can resolve a given intensity range – e.g. 256 grey values for a 8-bit camera or 64000 grey values for a 16-bit camera. The images can be shown on a computer monitor and saved on a computer - by photomultiplier tubes (PMT‘s): used often for scanning devices such as confocal laser scanning microscopes. The gain (voltage) of the PMT defines the sensitivity (electrons released for a given number of photons that hit the detector). Often more „noisy“ than CCD camera images. Averaging is used to smooth the images (good images takes about 4 sec – while CCD require just about 100 msec). - old fashioned: film camera and sensitive film (e.g. 1600 ASA) 217 108 Example for a fluorescence microscopy experiment - Cells transfected with fluorescent fusion proteins of a transcription factor and its inhibitor (appear in the cytosol); - addition of leptomycin B (LMB) to block nuclear export. This leads to accumulation in the nucleus indicating continuous nucleo-cytoplasmic shuttling 218 Example for a fluorescence microscopy experiment II Fluorescence was quantified in the nucleus and in the cytosol of the same cell after different time points > shows the kinetics of nuclear accumulation by the change of the cytosolic/nuclear ratio. cytosolic/nuclear fluor. The data were fitted by nonlinear regression analysis (single exp. decay) – leading to the half time of the nuclear import process. 5 4 3 2 1 0 0 20 40 min 60 80 219 109 Protocol of an immunofluorescence staining • • • • • • • • Fixation: 15 min 4% Paraformaldehyd 3x 5 min mit TBST wash (50mM TrisHCl pH7.4, 150 mM NaCl, 0.1%Triton) Block: 1 h at RT with 3% BSA in TBS Incubation with 1. Ab: anti-IκB (rabbit polyclonal, sc-371 Santa Cruz) 1:300 in TBS/3% BSA, over night at 4°C (or 1 h at 37°C). 2x 5 min wash with TBST, 1x 5 min with TBS Incubation with Alexa488 goat antirabbit 1:2000 in TBS/BSA: 1 h at 37°C 3x 5 min wash with TBST, 1x 5 min with TBS Mounting 220 Combinations of transmitted light and fluorescence IP-Lab Software A) direct acquisition with both light sources ImageJ software B) Separate acquisition of fluorescence and phase contrast and merge or blending (e.g. with ImageJ or other software) 221 110 Interactive Microscopy Demonstrations Very recommendable: http://micro.magnet.fsu.edu/ 1. Optical resolution: http://www.microscopyu.com/tutorials/java/imageformation/airyna/index.html http://www.microscopyu.com/tutorials/java/lightandcolor/refraction/index.html 2. Köhler Illumination: http://www.microscopyu.com/tutorials/java/kohler/index.html 3. Phase shift of light by an object http://www.microscopyu.com/tutorials/java/phasecontrast/phasespecimens/index.html 4. Phase contrast http://www.microscopyu.com/tutorials/java/phasecontrast/opticaltrain/index.html http://www.microscopyu.com/tutorials/java/phasecontrast/microscopealignment/index.html 5. Objectives with adjustable working distance http://www.microscopyu.com/tutorials/java/aberrations/correctioncollar/index.html 222 Confocal Laser Scanning Microscopy (CLSM) Problem in conventional microscopy: light, which comes from outside of the focal plane (above or below) gets to the detector (or eye) and is registered as blur, which decreases the quality of the image Solution: A pinhole (iris) is placed into the light path at a position, where it can block out-offocus light. By that means an optical section is imaged (with variable thickness starting with approx. 0.8 µm) depending on the diameter of the pinhole. Usually high quality excitation light is needed for that (e.g. coherent laser light with parallel light beams). The result is a very sharp image without any blur from out-of-focus light with a slightly higher resolution than with conventional epifluorescence microscopy. Photomultiplier confocal pinhole Laser dichroic mirror Scanner Objective z-Motor 223 111 Confocal microscopy removes the blur from thicker objects http://zeiss-campus.magnet.fsu.edu/tutorials/opticalsectioning/confocalwidefield/index.html Optical sectioning and 3Dprojections z-stack Acquisition of a „z-stack“ (image slices along the zaxis) allows reconstruction of a 3D-projection, which can be shown as animation projection 3D rendering 225 112 Spectral imaging Resolving spectral information on a pixel-by-pixel basis • • • • • • „Emission finger printing“: emission scan of a microscopy sample („lambda stack“ of images) at a given excitation wavelength (e.g. with Zeiss LSM META systems or with Leica confocal microscopes…) Alternative: Excitation scan (at a constant emission wavelength; e.g. using a monochromator light source) Combinations of excitation and emission finger printing (e.g using filter wheels) Increases the number of markers to be measured in parallel Can be used to discriminate fluorophores with overlapping spectra Can be used to discriminate specific fluorescence from autofluorescence Leica concept Zeiss META concept 226 Spectral Imaging Confocal Microscopy (with Emission Curve Analysis) Leica Confocal Microscope TCS SP2: Monochromator in front of the detector AOBS: Acousto-Optical Beam Splitter (instead of dicroic mirror) META System of Zeiss: 32 PMT-detectors every 10.7 nm (400 – 720 nm): simultaneous wavelength analysis. lambda-stack Spectral curve of a region of interest 227 113 Zimmermann et al. (FEBS Letters 2003) Sample with overlapping fluorophores 1 2 3 4 5 6 7 8 1 2 Emission curves separated into 8 channels (left) or 2 channels (right) Equation matrix for the channel signals based on reference intensities in the channels (GFPn and YFPn) and the unknown contributions of the fluorophores Unmixed fluorescence (pseudo-coloured) Spectral imaging example I: CFP, GFP and YFP http://zeiss-campus.magnet.fsu.edu/articles/spectralimaging/introduction.html 229 114 Spectral imaging example II: strongly overlapping dyes SYTOX Green (nucleus), Alexa Fluor 488 conjugated to phalloidin (filamentous actin network), and Oregon Green 514 conjugated to goat anti-mouse primary antibodies (targeting mitochondria). Invitrogen Spectra Viewer http://www.invitrogen.com/site/us/en/ home/Products-andServices/Applications/CellAnalysis/LabelingChemistry/FluorescenceSpectraViewer.html 230 Separation of specific fluorescence from autofluorescence by spectral imaging 231 115 Example for Emission Fingerprinting on a Zeiss LSM510 META: Separation of GFP and YFP Acquisition of a reference lambda stack for the first fluorophore (GFP) Obtain the spectral emission curve for the first fluorophore and repeat the procedure for the second fluorophore Intensity 250 YFP 200 GFP 150 100 50 0 500 510 520 530 540 550 560 Emission wavelength (nm) 116 Unmixing of a mixed sample (GFP-Actin and YFP-membranes) Emission stack Unmixed image Unmixing of signals in pathology samples (Shown with the Nuance™-Software from Cambridge Research & Instrumentation) image with mixed signals for different markers autofluorescence Brightfield display 235 117 „Realtime“ confocal microscopy, Spinning disk confocal microscopy (with Nipkow-disks) gentle scanning (less bleaching > good for sensitive life cells Detection of the signal with a CCDcamera http://zeiss-campus.magnet.fsu.edu/tutorials/spinningdisk/yokogawa/index.html 236 Companies for confocal microscopes • Zeiss: http://www.zeiss.de • Leica: http://www.leica.com www.confocal-microscopy.com • Nikon: http://www.instrumente.nikon.de/ • Olympus: http://www.olympus.de/microscopy/ 237 118 Multiphoton Laser Scanning-Microscopy A quantum physical phenomenon is used: at very high light densities (using pulsed lasers, about 900 nm infrared light) packages of 2 or more photons occur (just in the focal plane !). These have the same energy as single photons of higher energy (shoerter wavelength, e.g. 450 nm). Thus these photon packages can excite a fluorophore, which emits then at for instance 520 nm (mitted wavelength is horter than the excitation light wavelength !). An important advantage is that the 900 nm light has a mucher deeper penetration into tissue (approx. 1 mm), while conventional excitation can image just down to 0.25 mm. Another advantage is a reduced overall bleaching effect, as excitation photon packages occur just in the focal plane. 238 Multiphoton Laser Scanning-Mikroskopie II conventional excitation (1-Photon > cone of ecitation light) 2-Photon excitation: only a spot of excitation 239 119 Special Fluorescence Microscopy Techniques 1. FRAP: Fluorescence Recovery After Photobleaching 2. FLIP: Fluorescence Loss in Photobleaching 3. FRET: Fluorescence Resonance Energy Transfer 4. FLIM: Fluorescence Lifetime Imaging Microscopy 5. FISH: Fluorescence In Situ Hybridization 240 FRAP: Fluorescence Recovery After Photobleaching FRAP at the membrane Non linear regression analysis y = span (1-e-kx) + bottom An image is taken – then a region of the cell is bleached by high laser intensity, followed by a time series of images after bleaching. Briefly after bleaching the region is significantly darker and then the fluorescence intensity increases again (fluorescence reoovery) due to diffusion of molecules into the bleached area. The kinetics of recovery depends on the diffusion coefficience; the extent of recovery (the plateau to which the fluorescence recovers) is a measure of the overall mobility (the fraction of mobile molecules versus molecules immobilized, e.g. to the cytoskeleton) 241 FRAP in the cytosol: 120 inverse FRAP with novel fluorescent proteins Protocol: FRAP analysis on Zeiss LSM510 • • • • • • • • Capture an image of the whole cell before bleaching Define a bleaching / scan region (and maybe in addition another scan region that is not bleached) Perform a time series with 1 scan prebleach, about 70 iterations of bleaching with 100% laser power and then 50-100 scans of the bleach region (and also the non-bleached control region if you specified one)- a good time resolution can only be obtained if just the small bleach region (and maybe the control region) is scanned - and not the whole cell; averaging of 2 or 4 scans reduces the electronic noise and leads to better quantifications. Capture an image of the whole cell after the FRAP time series (with the same conditions as the prebleach image – for calculating the total loss of fluorescence. If you want to save disk space: extract the FRAP region and save just this region instead of the whole image It is recommended to use the WCIF version of ImageJ for analysis: You can open the LSMfiles with the built-in feature (which also allows opening the time values of the image series). Measure the mean fluorescence in a control region or for the whole cell for both the prebleach and the postbleach images and calculate the loss of overall fluorescence due to the bleaching in the region of interest (this is necessary for obtaining correct recovery values for the bleach region). Import the FRAP-image sequence, define a measurement region and apply the „intensity versus time plot“ plug-in – this will draw a graph of the FRAP curve; clicking the list button, shows a list of the numerical values (the first 4 parameters are dimension and position of the region, the rest are the fluorescence intensity values). Copy the fluorescence raw data from the list to the corresponding column of an Excel template 243 121 • Calculate the difference of mean fluorescence from the background and normalize the fluorescence values to 100% for the initial fluorescence. • Divide the percent values by the correction factor calculated from the total loss of fluorescence (e.g. if total fluorescence decreased from 1 to 0.9 then divide the mean fluorescence of the FRAP regions for each time value by 0.9 to compensate for the loss in total fluorescence). A similar compensation can be obtained by normalizing the FRAP fluorescence values to the control scan region that was not bleached. This method also compensates more exactly for the bleaching effect in the course of scanning of the time series (this scanning-dependent bleaching effect is opposed to the recovery of fluorescence in the bleach region due to diffusion of non-bleached molecules in the bleach region). This “dynamic correction” gives a somewhat better estimation of the curve (and the kinetics of the recovery) – but leads in principle to results that are very similar to the curve obtained with the “constant correction factor” (by calculating the total loss in fluorescence based on the intensities of the images that were captured before and after the FRAP-time series) • For non-linear regression analysis (curve fit of the data to a single exponential association algorithm): Copy the data to a fitting program (such as GraphPad Prism) and perform the fitting with a “bottom to span” algorithm: y = span × (1 − e− kx ) + bottom 244 FLIP: Fluorescence Loss in Photobleaching … to determine the dynamic shuttling of molecules between different compartments of the cell A certain compartment A (e.g. the cytosol) is repetitively bleached by the laser – and the fluorescence decrease in a different compartment B is monitored by time lapse microscopy. Molecules that shuttle from B to A are bleached in A > thus the compartment B gets dimmer when there is a dynamic distribution of molecules between A and B. 120 cytosol 100 nucleus 80 60 40 20 0 0 2 4 6 122 FLIP to determine a nuclear export signal and a nucleolar localization signal NFκB inducing kinase truncated NIK without the export sequence: nuclear FLIP (bleach in nucleus outside nucleoli) 125 nuclear nucleolar 100 75 50 25 0 0 100 200 300 400 500 600 700 sec FCS: Fluorescence Correlation Spectroscopy … to determine diffusion coefficients and interactions between molecules. The sample is illuminated by the laser at a very small spot, the movements of molecules in this spot (in and out) cause fluorescence fluctuations, which are analyzed by correlation functions 123 FRET: Fluorescence Resonance Energy Transfer Microscopy Energy can be transferred between two fluorophores when they are very close to each other (closer than 10 nm) and when the emission curve of one (the energy donor) overlaps with the excitation curve of the other one (the acceptor). This transfer of energy does not happen via photons (!) but by a dipole-interaction (a quantum physical phenomenon discovered by Theodor Förster in 1946). As a result the donor fluorophore fluorescence becomes weaker and the acceptor fluorescence increases. The FRET effect decreases with the 6th power of the distance; the distance of half maximal energy transfer is called Förster-Distance R0 (for CFP and YFP it is approximately 5 nm). As the effect is usually not detectable anymore at a distance higher than 10 nm it is ideally suited for monitoring macromolecular interactions (protein-protein or protein-DNA). By that means, not only the interaction by itself can be detected, but also the location of the interaction and its dynamics. 248 Principle of Fluorescence Resonance Energy Transfer no FRET Donor FRET Acceptor donor fluor. (CFP) acceptor fluor. (YFP) E = R06/(R06 + r6) und nm excitation excitation emission Donor Acceptor R0 = [κ2 × J(λ) × n-4 × Q]1/6 × 970 R0 … Förster-Distance r ….. real distance κ ….. Orientation factor J(λ) … spectral overlap n … refractive index Q … Quantum yield of fluor. 249 124 Appropriate Fluorophore Pairs for FRET Fluorophore Pair Comments CFP / YFP Good combination for normal FRET microscopy using Hglamps as light source and special filters. CFP is poorly excitated by Ar-lasers, but: good excitation by blue laser diodes BFP / GFP BFP has inferior fluorescence properties GFP / DsRed-variants Original DsRed is just fluorescent as tetramer, shows complex maturation characteristics with green fluorescent intermediates and tends to aggregate; DsRed2 is a dimer. The new monomeric DsRed works very fine with GFP. GFP / YFP Are very difficult to separate with filters (but can be used in FLIM and with spectral analysis) GFP / Cy3 or Alexa 546 Antibodies can be directly labeled with Cy3 or equivalent Alexa dye and give FRET with a GFP chimera to which they bind. FITC / TRITC Classical FRET pair for labeled proteins (e.g. antibodies) Alexa 488 / Alexa 546 (Cy3 / Cy5) alternatives as labeling dyes (superior to FITC and TRITC) 250 FRET can be used to monitor protein-DNA interactions spectra of donor and acceptor (GFP-NF-κB and Tet-labeled DNA, respectively) 1.2 GFP-NFkB (Em) Tet (Em) 1.0 relative fluorescence Spektral analysis of a mixture: Increase in acceptor fluorescence indicates FRET Tet (Ex) 0.8 0.6 0.4 0.2 Spectral overlap 0.0 480 500 520 540 560 580 600 nm Excitation of GFP (488 nm) GFP-Emission (512 nm) GFP GFP-NF-κB DNA Tet-Emission (540 nm) FRET Tet-label 251 125 FRET can be applied to visualize the interaction of signaling molecules in living cells decrease in Donor Emission ECFP and EYFP-Scans 1,2 relative fluorescence EYFP (Em) 1,0 increase in Acceptor Emission EYFP (Ex) ECFP (Em) 0,8 ECFP (Ex) 0,6 FRET 0,4 0,2 0,0 350 Spectral overlap 1 nm 400 450 500 550 600 nm Förster Distanz R0 for ECFP and EYFP: ca. 5 nm (50% FRET Effizienz): > no FRET-Signal beyond 10 nm. X Y Donor Acceptor 252 Overview of FRET-Microscopy Techniques 1. Acquisition with FRET filter set (donor excitation and acceptor emission): Problem: coexcitation of the acceptor at the donor wavelength > false positives 2. Acquisition of a ratio image of acceptor fluorescence at donor excitation and donor fluorescence at donor excitation 2-Filter FRET Microscopy: Just works when there is equal expression of donor and acceptor (e.g. in fusion protein, biosensors) 3. 3-Filter FRET Microscopy 4. Determine the kinetics of donor fluorescence bleaching (this is slower in the presence of a FRET acceptor, as part of the bleaching energy is transfered to the acceptor) 5. Donor recovery after acceptor photobleaching 6. fluorescence lifetime microscopy– FLIM; fluorescence lifetime of the donor decreases in the presence of a FRET acceptor 253 126 Spectral crosstalk of donor and acceptor ECFP and EYFP-Scans 1.2 relative fluorescence 1.0 raw FRET-channel: 0.8 Donor Excitation + Acceptor Emission 0.6 0.4 0.2 0.0 340 360 380 400 420 440 460 480 nm Excitation window of donor 500 520 540 560 580 600 Emission window of acceptor Problems: 1. Co-excitation of the acceptor at the Donor-excitation wavelength > Non-FRET-Fluorescence in the raw-FRET channel 2. Signal-overlap of donor into the acceptor channel > Non-FRET fluorescence in the raw-FRET channel 254 2 Filter-FRET Microscopy (Ratio Imaging) Ratio of donor emission and acceptor emission at the excitation wavelength of the donor Limitations: •concentration dependent •donor and acceptor have to colocalize completely ECFP and EYFP-Scans 1.2 EYFP (Em) EYFP (Ex) relative fluorescence 1.0 ECFP (Em) ECFP (Ex) 0.8 0.6 0.4 0.2 0.0 350 400 450 500 550 600 nm just useful for FRETbiosensors with covalent linkage between donor and acceptor (equal-molar expression and 100% colocalization) excitation emission-1 emission-2 image = Emission2 : Emission1 255 127 3-Filter FRET Microscopy 3 Images are taken (under constant camera settings): 1. Donor (e.g. CFP-excitation and emission), 2. Acceptor (e.g.YFP-excitation and emission – this signal is not affected by FRET 3. FRET-Filter (raw FRET: CFP-excitation and YFP-emission). A normalized FRET signal (image) can be calculated by using correction factors obtained with single stained samples: FRETc = IFRET - corrCFP x ICFP – corrYFP x IYFP corrCFP : ca. 0.59 corrYFP : ca. 0.18 CFP / YFP neg. control CFP-YFP pos. control 256 FRET microscopy example corrected FRET = IFRET - corrCFP x ICFP – corrYFP x IYFP sample Donor channel Acceptor channel FRET channel corr. factor corrected FRET CFP alone 100 0 60 0.6 0 YFP alone 0 100 20 0.2 0 non-bound CFP + YFP 100 100 80 0 bound CFP-YFP 100 100 160 80 Donor neg. control Acceptor corrected FRET normalized FRET Normalized FRET (normalized to diff. expression levels): = × sample 257 128 FRET Microscopy by analyzing the kinetics of donor bleaching … this is slowed down in presence of a FRET acceptor time series of images time series of images 436 nm ECFP FRET 476 nm EYFP 436 nm 476 nm ECFP CFP-Protein alone CFP- and YFP-Protein 258 Donor-bleaching kinetics Probe mit FRET advantages: concentration independent donor and acceptor don‘t have to colocalize completely Limitation: requires external control, difficult to obtain a FRET-image single exponential decay y = A 0 .e −kt + offset Probe ohne FRET y... Fluor. Signal A0... starting signal k... decay constant t... time offset... final value Fluorescence half time Tau: τ = 0.69/k FRETeff. E = 1 - (τ without / τ with Akzeptor.) 259 129 FRET Microscopy by acceptor bleaching and monitoring donor recovery (do not use for CFP / YFP) Donor recovery after acceptor bleaching: An image of the donor in the presence of the acceptor is taken, then the acceptor is bleached (partially), followed by acquisition of a second donor image Donor Donor FRET Acceptor Acceptor 260 Visualisation of biochemical reactions by FRET microscopy (e.g. phosphorylations) Detection of the auto-phosphorylation of EGF-receptor on Tyrosine residues using GFP-EGFR fusion protein and Cy3-labeled anti-P-Tyr antibodies (donor recovery after acceptor photobleaching technique) erb2 -P GFP Cy3 GFP Ratio image 261 130 FLIM: Fluorescence Lifetime Imaging Microscopy The lifetime of donor fluorescence (usually in the nanosec. range) is reduced in presence of a FRET acceptor. This lifetime can be determined by a special variant of microscopy. Usually a pulsed or a modulated laser is used for excitation. The fluorescence decay (Time Domain) or the phase shift (Frequency Domain) of the emission compared to the excitation is a measure of the fluorescence lifetime. fluor. image FLIM image 262 FRET-Biosensors I Caspase 3-Biosensor: Apoptosis (Activation of caspase 3) is detected with a CFP-YFP fusion protein in which CFP and YFP are separated by a caspase 3 cleavage site. Without apoptosis: FRET, with apoptosis: no FRET CFP Caspase 3 -…DEVD…- YFP FRET no FRET 263 131 Other examples for FRET-Biosensors Calcium-Biosensor: Ca2+-sensitive Calmodulin and a Ca2+/Calmodulin-binding M13 domain are spliced between CFP and YFP (additional localization sequences can be added – e.g. signal peptide and ER retention sequence). A change in the calcium concentration leads to a change in the conformation of the linker and thus to an alteration of the FRET signal. Ca2+ YFP YFP CFP FRET CFP Calmodulin M13 Ca2+ PKA Activity sensor: CFP and YFP separated by a PKA-substrate sequence and a 14-3-3 domain, which binds phospho-serine of the PKA substrate domain. FRET PKA-substrate 14-3-3 CFP YFP PKA CFP YFP P 264 Digital Image Analyses Often used: ImageJ (scientific Freeware: http://rsbweb.nih.gov/ij/ ) Different operations can be performed: contrast enhancement, smoothing, background subtraction, measurement of fluorescence intensities… – and further more even math with images can be done (e.g. dividing one image by another one; each pixel value is divided by the corresponding pixel value of the second picture – at the same pixel coordinates) 265 132 Division of images with ImageJ 266 PixFRET Plugin for ImageJ 133 FRET analysis with self-written ImageJ macro neg. control 14 12 10 8 6 4 2 0 Negative Control IKK1+Myc IKK2+ Myc sample 1 sample 2 sample In Situ Hybridisation (ISH) … for specific detection of DNA or mRNA sequences. A labeled DNA- or RNA is hybridized to the target sequence in situ (in the cell or the tissue) and detected Applications: 1. Detection or (semi-) quantification of mRNA 2. Detection of DNA-sequences in chromosomes (e.g. translocations, mutations, loss of genes…) http://www.cytochemistry.net/In_situ.htm http://osiris.rutgers.edu/~smm/in_situ_hybridization.htm 269 134 Principle of ISH - a labeled“ probe (e.g. DNA, labeled with Biotin-dUTP by Nick-Translation or an oligonucleotide, labeled by terminal deoxynucleotidyl transferase, TdT) diffuses into the cell and hybridizes with the target sequence. Addition of formamide to the hybridization buffer lowers the specific hybridization temperature, so that at 37°C only specific target sequences are bound. The probe is then detected via fluorescence (fluorescence in situ hybr., FISH) or by enzyme activity (e.g. HRP and colour reaction). FITC 270 ISH: Advantages and Disadvantages of various probes Probe type Advantages Disadvantages DNA (double strand) Easy to use Subcloning unnecessary Choice of labeling methods High specific activity Possibility of signal amplification (networking) Reannealing during hybridization (decreased probe availability) Probe denaturation required, increasing probe length and decreasing tissue penetration Hybrids less stable than RNA probes DNA (single strand) No probe denaturation needed No reannealing during hybridization (single strand) Technically complex Subcloning required Hybrids less stable than RNA probes RNA Stable hybrids (RNA-RNA) High specific activity No probe denaturation needed No reannealing Unhybridized probe enzymatically destroyed, sparing hybrid Subcloning needed Less tissue penetration Oligonucleotide No cloning or molecular biology expertise required Stable Good tissue penetration (small size) Constructed according to recipe from amino acid data No self-hybridization Limited labeling methods Lower specific activity, so less sensitive Dependent on published sequences Less stable hybrids Access to DNA synthesizer needed from: Feldman, RS, Meyer, JS, and Quenzer, LF (1997). Principles of Neuropsychopharmacology. Sunderland, MA: Sinauer Associates, Inc. Pages 31-35. more recently: BAC probes (can be obtained from collections): high sensitivity for single copy genes… 271 135 Examples for ISH FISH with interphase nuclei Fluorescence In Situ-Hybridization on Metaphase-chromosomes Detection of a target-mRNA in cryoor paraffin sections Detection with 33P Detection with alkal. Phosphatase 272 mRNA-FISH Protocol • • • • • • • • • • • • Cells are fixed with freshly made 4% formaldehyde in PBS, pH 7.4 for 15 min at room temperature. All solutions should be made in Molecular Biology grade ultrapure water (no RNase). Wear gloves at all times and use sterile disposable pipets and tips. After rinsing in PBS (3 X 10 min. each), cells are permeabilized with 0.5% Triton X-100 in 1X PBS for 10 min at 4oC. Cells are then rinsed in PBS (3 X 10 min. each) and then in 2X SSC (1 X 5 min.). 100 ng of nick translated probe (containing digoxigenin dUTP) and 20 ug of competitor E. coli tRNA per coverslip are dried down in a Speed Vac (Savant). This is a good starting place but you may have to titrate your specific probe. 10 μl of deionized formamide is added to the dried DNA. The probe and tRNA are denatured by heating for 10 min at 90oC. The probe is chilled on ice immediately. 10 μl of Hybridization buffer (20% dextran sulfate + 4X SSC) is added to the denatured probe so that the final concentrations in the hybridization mixture are 5 ng/ml of probe, 1 ug/ml of E. coli tRNA, 2X SSC, and 10% dextran sulfate. 20 μl of hybridization mixture/probe is placed onto each coverslip. Coverslips are inverted onto a slide and sealed with rubber cement and incubated in a humid chamber for 16 hrs. at 37oC. After rinsing in 2X SSC/50% formamide at 37oC, 2X SSC and 1X SSC at room temperature for 30 min. each, the coverslip containing cells are incubated in 4X SSC/0.25% BSA/2ug/ml anti-digoxigenin antibody for 60 min. in a humid chamber at room temperature in the dark. Coverslips are then rinsed in 4X SSC (1 X 15 min.) at room temperature, 4X SSC/0.1% Triton X-100 (1 X 15 min.), and 4X SSC (3 X 10 min. each). Coverslips are mounted in fluorescence mounting medium. Modified from: Jiménez-García, L. and D.L. Spector. 1993. Cell 73, 47-59. 273 136 Superresolution Microscopy I STED: Stimulated Emission Depletion A second laser (depletion laser) „trims“ the excitation spot (point-spread function, PSF) to a smaller size. Resolution appr. 80 nm. http://zeiss-campus.magnet.fsu.edu/articles/superresolution/introduction.html 274 Superresolution Microscopy II Structured Illumination Microscopy (SIM) A second laser (depletion laser) „trims“ the excitation spot (point-spread function, PSF) to a smaller size. Resolution appr. 80 - 100 nm. normal image SIM image A known pattern is projected into the image plane at different angles and interferes with sample structures, creating Moiré pattern. Superresolution information can now be captured by the microscope from these structures by mathematical algorithms. 275 (from www.zeiss.de) 137 Superresolution Microscopy - by single molecule detection STORM: Stochastic Optical Reconstruction Microscopy using single fluorescent molecules PALM: Photoactivated Localization Microscopy Resolution: appr. 30 nm, based on statistical calculation of the center of a Gaussian Fit of a single molecule. Requires a sensitive camera (e.g. EMCCD: Electron-multiplying chargecoupled device cameras) – and some software, but no specific hardware http://zeiss-campus.magnet.fsu.edu/articles/superresolution/introduction.html 276 Research Methods - Overview • • • • • • • • cell culture systems labelling and transfection of cells analyses of cellular components analyses of molecular interactions fluorescence measurements microscopy flow analysis (FACS) analyses of cellular processes (proliferation, apoptosis..) 277 138 Flow Cytometry Some contents are inspired by “Fluorescence Spectroscopy in Biological Research” by Robert F. Murphy Definitions: • Flow Cytometry – Measuring properties of cells in flow • Flow Sorting – Sorting (separating) cells based on properties measured in flow – Also called Fluorescence-Activated Cell Sorting (FACS) 278 Basics of Flow Cytometry •Cells in suspension Fluidics •flow in single-file through •an illuminated volume where they Optics •scatter light and emit fluorescence •that is collected, filtered and Electronics •converted to digital values •that are stored on a computer - Flow Cytometry, Flow Analysis). - Flow Sorting, Fluorescence Activated Cell Sorting, FACS 279 139 http://probes.invitrogen.com/resources/education/ 280 Fluidics • Need to have cells in suspension flow in single file through an illuminated volume • In most instruments, accomplished by injecting sample into a sheath fluid as it passes through a small (50-300 µm) orifice 281 140 Fluidics II • When conditions are right, sample fluid flows in a central core that does not mix with the sheath fluid • This is termed Laminar flow • Whether flow will be laminar can be determined from the Reynolds number V is the mean fluid velocity in (SI units: m/s) D is the diameter (m) μ is the dynamic viscosity of the fluid (Pa·s or N·s/m²) When Re < 2300, flow is always laminar ν is the kinematic viscosity (ν = μ / ρ) (m²/s) When Re > 2300, flow can be turbulent Q is the volumetric flow rate (m³/s) ρ is the density of the fluid (kg/m³) A is the pipe cross-sectional area (m²) 282 Flow Cell Injector Tip Fluorescence signals Sheath fluid Hydrodynamic Focusing Focused laser beam 283 Purdue University Cytometry Laboratories 141 Hydrodynamic Focusing The figure shows the mapping between the flow lines outside and inside of a narrow tube as fluid undergoes laminar flow (from left to right). The fluid passing through cross section A outside the tube is focused to cross section a inside. 284 V. Kachel, H. Fellner-Feldegg & E. Menke - MLM Chapt. 3 Hydrodynamic Focusing II Example: Focusing of ink by laminar flow into a capillary Notice how the ink is focused into a tight stream as it is drawn into the tube under laminar flow conditions. In flow cytometry: Sample container and the sheath fluid container are put under defined air pressure. Laminar flow is maintained (and hydrodynamic focussing is achieved) by a precise control of the pressure difference between the sample container, the sheath fluid container and the atmosphere 285 V. Kachel, H. Fellner-Feldegg & E. Menke - MLM Chapt. 3 142 Flow Chamber Different types are possible, a commonly used type: Flow through cuvette (sense in quartz) 286 H.B. Steen - MLM Chapt. 2 Optics: General scheme PMT 4 Flow cell PMT Dichroic Filters 3 PMT 2 Bandpass Filters PMT 1 FSC Laser SSC 287 original from Purdue University Cytometry Laboratories; modified by R.F. Murphy and J. Schmid 143 Forward Scatter (FSC) When a laser light source is used, the amount of light scattered in the forward direction (along the same axis that the laser light is traveling) is detected in the forward scatter channel The intensity of forward scatter is mainly proportional to the size and surface properties of cells (or other particles) 288 Forward Scatter Detector Usually not a sensitive PMT detector (because the light intensity is high) – but rather a photodiode detector, which can be set in log10increments (E-1, E0, E1...), and a linearity gain factor (1.0 – 9.99). A blocking bar prevents the direct laser light from hitting the detector. A predefined FSC-Wert is often used as threshold to discriminate between cells and dust particles 289 144 Forward scatter threshold to ignore debris and small particles or cells 290 • When a laser light source is used, the amount of light scattered to the side (perpendicular to the axis that the laser light is traveling) is detected in the side or 90o scatter channel • The intensity of side scatter is mainly dependent on subcellular structures (e.g. granules, vesicles…) Side Scatter (SSC) 291 145 90 Degree Light Scatter (SSC) Laser FSC Detector 90° scattered light (side scatter) is detected with a photomultiplier detector, where the sensitivity can be set in Volt. Amplification scale can be linear or logarithmic. SSC detector 292 Purdue University Cytometry Laboratories Optics: Fluorescence Channels dichroic mirrors bandpass filters 146 Common Laser Lines 350 300 nm 457 488 514 400 nm 500 nm 610 632 600 nm 700 nm Fluorophores PE-TR Conj. Texas Red PI Ethidium PE FITC cis-Parinaric acid 294 Purdue University Cytometry Laboratories Sorting of cells FL1 488 nm Laser FSC Sensor Fluorescence - FSC charged electrodes - + + - Gating control unit Sorted cells 295 Purdue University Cytometry Laboratories; modified 147 296 Sorting Mode If there are 2 or more cells in one droplet, and the droplet contains target and non target cells, then there is a „sorting conflict“, which has to be solved by defining an appropriate sorting mode: Target-Cell Non-target cell • Exclusion-Mode: droplets containing non target cells are dismissed even if they contain target cells > high purity, maybe lower yield • Recovery-Mode: droplets containing target cells are collected, even if they also contain non-target cells > high yield, maybe low purity. • Single Cell Mode: only droplets with single target cells are collected > high purity, maybe low yield, high counting accuracy. 297 148 Overlap of fluorescence signals and „Compensation“ 298 „Compensation“ When cells are labeled with 2 fluorophores (e.g. FITC and PE), there might be a signal crosstalk between the detection channels (dependent on detector voltage settings). This can be compensated in the follwing way: measuring a sample containing only fluorophore 1: if there is crosstalk, you see an elevated intensity in both channels – using the compensation control of the software, you can bring the fluorescence signal in the wrong channel down to the background fluorescence. With a sample stained only with fluorophore 2 you do the same for the other channel. This compensation is usually just valid for the detector settings at which it was set. compensation for fluorophore 2 299 compensation for fluorophore 1 149 Compensation II before compensation after compensation > true double positives can be determined 300 FACS-Graphs counts Histograms: the intensity of one channel is divided into classes (often 1024, x-axis) and the frequency of „events“ (cells) scored into these 1024 classes. 0 FL1 Dot Plots: 2 different parameters are plotted on the x- and y-axis; each cell is a spot on this graph according to its parameter intensities. FL2 Correlation between histograms and dotplots 301 150 FACS-Graphs II FL1 Density Plots: The frequency of events (cells) in various areas of a dot plot is colour coded to highlight the peaks Contour Plots: The frequency of events (cells) in various areas of a 2D-plot is visualized by lines representing equal probability (similar to contour lines of mountains on a map) FL2 FL1 FL2 3D-Plots: The frequency of events measured for 2 parameters is illustrated by a third axis (z-axis) in a 3D manner. FL2 302 Statistics and „Gating“ Region R1: 69.3% Regions: can be defined in different forms (polygons, ellipses..). These can be used for statistics of cells falling into a certain region – but also for „gating“ that means rejecting certain cells inside (or outside) a certain region for analysis (or data acquisition) – or for instance for sorting of cells. (Polygon-Region) Marker M1: 67.5% Marker: Upper and lower limits in histograms for quantification Quadrants: split the 2D graph in 4 regions, by the coordinates of the point where 2 axis intersect. Allows fast and simple statistics. UL (upper left): 0.06% UR (upper right): 29. 88% LL (lower left): 0.64% LR (lower right): 69.44% 303 151 Research Methods - Overview • • • • • • • • cell culture systems labelling and transfection of cells analyses of cellular components analyses of molecular interactions fluorescence measurements microscopy flow analysis (FACS) analyses of cellular processes (proliferation, apoptosis..) 304 Analytical Applications of Flow Analysis • • • • • • • • • Leukocyte analyses Phenotyping of cells (CD-Marker) Immunofluorescence stainings cell cycle analyses (DNA-content) Chromosome analyses Proliferation assays (Brd-U incorporation) Apopotosis assays (Annexin V, TdT, JC1) Calcium flux-measurements etc. 305 152 Leukocyte analyses Phenotyping of cells CD8-positive T-cells CD4-positive T-cells 306 cell cycle analyses by staining of DNA with PI diploid Chromosome content G2M G0/G1 s doubled diploid G0 G1 G0G1 apoptotic cells G2/M S s 0 Propidium-Iodide Fluorescence of permeabilized cells after digestion of RNA: fluor. depends on DNA content 200 400 G2 M 600 4N 2N DNA Gehalt 800 1000 307 153 Protocol of a Propidium-Iodide Staining Adherent cells: trypsinized, suspended in medium + 10% FCS, centrifuged (1000 rpm, 5 min), Pellet suspended in PBS (1 ml) Suspension cells: Centrifuged (1000 rpm, 5 min), Pellet suspended in PBS (1 ml) Fixation with EtOH: Pipet cell suspension into 2.5 ml absolute EtOH (final concentration approx. 70%) - or vortex the suspension at half speed while adding the EtOH) – to prevent clustering of cells during the fixation. Incubate on ice for 15 min (or over night at –20°C). Alternative fixation with paraformaldehyde: Pipet the 1 ml cell suspension into 3 ml 4% paraformaldehyde and fix for 15 min at r.t. Staining: Pellet the cells at 1500 rpm for 5 min, Suspend the pellet in 500 µl PI-solution in PBS: 50 µg/ml PI from 50x stock solution (2.5 mg/ml), 0.1 mg/ml RNase A, 0.05% Tritin X-100 Incubate for 40 min at 37°C Add 3 ml of PBS, pellet the cells (1500 rpm, 5 min) and take off the supernatant Suspend the pellet in 500 µl PBS for flow analysis (you can also leave about 500 µl of the diluted staining solution on the pellet and suspend the cells in this solution > less loss of cells when you take off the sup.) – the rest of the staining solution does not interfere with the flow analysis. Flow analysis: Approximate settings (on FACSort): FL1: 570 V log. (e.g. if you want to detect GFP) FL2: 470 V linear 308 Chromosome Analyses 309 154 Analysis of Proliferation (BrdU-labeling of S-Phase Cells) Cells are cultured for a given time in medium containing Bromodeoxy-Uridin (BrdU). This nucleotide analogon incororates into newly synthesized DNA (of cells in S-phase) – and can be detected with anti-BrdU antibodies (e.g. FITC labeled). FITC-antiBrdU Fluor. Propidiumjodid Fluor. (PI) 310 Staining of apoptotic cells with JC-1 JC-1 (5, 5´, 6, 6´-tetrachloro-1, 1´, 3, 3´-tetraethylbenzimidazol-carbocyanine iodide) is a dye, which incorporates in to mitochondrial membranes, where the fluorescence depends on the membrane potential. In normal cells (with intact mitochondrial membrane potential) it builds aggregates and emits mainly red fluorescence, in apoptotic cells (where the membrane potential breaks down) it occurs in monomers and emits mainly green fluorescence. This can be detected by flow analysis or microscopy. 311 155 Protocol of a JC-1 Staining of Apoptotic Cells JC-1 is prepared as a 1000x stock solution in DMSO (5 mg/ml). For the staining of adherent cells it is diluted in medium to 5 µg/ml (with vortexing during the dilution to prevent the formation of precipitates); the JC1 containing medium is added to the cells, followed by incubation for 10 min at 37°C (or RT for 15 min). Subsequently the cells are washed twice with PBS, trypsinized, suspended in 500 µl PBS and analyzed by flow analysis. Suspension cells (lymphocytes): suspend 1:1 with 10 µg/ml JC-1 in medium (final conc.: 5 µg/ml) Approximate detection settings on FACSort: FL1: 360 V (log) FL2: 310 V (log) Compensation : FL1-7% FL2 und FL2-74% FL1 312 Detection of apoptotic cells by PI-staining of permeabilized cells (cell cycle analysis) Fragmented DNA emits lower fluorescence then cells with the normal diploid DNA content – this „Sub-G0/G1“ population reflects apoptotic cells with fragmented DNA )at a late stage of apoptosis) 313 156 Detection of apoptotic cells with Annexin V Fluorescence-labeled Annexin V: binds to phosphatidylserine, which is normally on the inner leaflet of the membrane, but which is flipped to the outside during apoptosis Discrimination between necrotic and apoptotic cells PI-pos/Annexin-neg.: necrotic PI-neg/Annexin-pos.: early apoptotic PI-pos/Annexin-pos.: late apoptotic, or necrotic with large holes in the membrane (where Annexin V can get through) 314 Calcium Flux Determination by Flow Analysis Cells are stained with a calcium sensitive fluorophore (INDO-1 or better Fluo4); after a stimulus, the kinetics of the fluorescence signal is measured (monitoring calcium influx). 315 157 Sources • Purdue Univ. Cytometry Laboratories http://www.cyto.purdue.edu/ • Research Institute of Scripps Clinic: http://facs.scripps.edu/ • Invitrogen Tutorials (http://www.invitrogen.com/site/us/en/home/support/Tutorials.html) • • • • BD Tutorials: http://www.bdbiosciences.com/support/training/itf_launch.jsp J. Paul Robinson (Purdue Univ.) Robert F. Murphy (Carnegie Mellon Univ., Pittsburgh) Flow Cytometry and Sorting, 2nd ed. (M.R. Melamed, T. Lindmo, M.L. Mendelsohn, eds.), Wiley-Liss, New York, 1990 – abgekürzt: MLM • Flow Cytometry: Instrumentation and Data Analysis (M.A. Van Dilla, P.N. Dean, O.D. Laerum, M.R. Melamed, eds.), Academic Press, London, 1985 – abgekürzt: VDLM 316 Companies offering flow analysis equipment • • • • • Becton-Dickinson http://www.bd.com Beckman-Coulter http://www.beckmancoulter.com Millipore: http://www.millipore.com/ Accuri (> now part of BD) http://www.accuricytometers.com/ Partec: http://www.partec.com/ 317 158 Free Software: Flowing Software 2 http://www.flowingsoftware.com 318 Flowing Software 2 Quadrants Regions in histograms 319 159 320 Cell cycle analysis with Flowing Software • Define 3 histogram regions (H1, H2, H3: G0/G1, S and G2/M-phase, respectively) • Activate the region control tool: Cell Cycle • Define the G2 peak multiplier and peak width (right click in the cell cycle window) • Choose active control (after right clicking) • Create statistics (by right clicking into the histogram window) • Create a Stat.List by right-clicking into the Statistics window • Ctrl-N loads the next file, adjusts the H1-H3 regions automatically and calculates the cell cycle phases (H1-H3) 321 160 Tissue Cytometry: Quantitative Image Analysis of microscopy samples Single-cell recognition (e.g. based on DAPI-nuclear fluorescence) > generation of cell masks for quantification of signals (e.g. in different fluorescence channels) > scattergrams can be derived similar to flow cytometry (for the cells in their original tissue environment!) > Gating and statistics are possible http://www.tissuegnostics.com Similar evaluations can be done with ImageJ using automatic threshold and the „Analyze particles“ feature. Tissue Arrays for quantitative comparison of samples > equal staining conditions for all samples CA Normal PIN Pat. #1 Pat. #2 Pat. #3 Pat. #4 Control Gingiva Cystectomy Example of a prostate tissue array 161 Multi-parallel coordinate plots for visualization of several parameters in parallel Can be done with Freeware (Mondrian: http://www.theusrus.de/Mondrian/index.html) One event (e.g. one cell) is represented by a line linking several y-axis (for the different parameters e.g. fluorescence signals); a „population“ can be selected and is highlighted also for the other parameters. The data density can be reduced (using a so called alphafactor) to obtain better visibility of numerous data points. 1 2 3 4 5 6 7 8 9 10 The density of lines (events) can be visualized in colourcoded manner (example: 10-parameter parallel coordinate plot generated with MatLab) > fast intuitive visualization of multiparameter data sets 162