Neuroanatomical Education Using L
advertisement

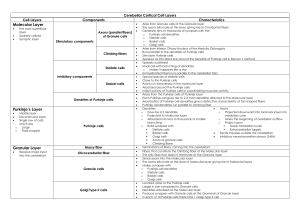
Neuroanatomical Education Using L-Systems: Modeling Neuronal and Glial Cells Tatiana Karaman Fall 2013 Tatiana Karaman ABSTRACT Neuroanatomy is an essential part of many kinds of biomedical research and education, but there is a clear lack of consistency in visual and numerical data in textbooks and online resources. This issue is abundantly clear when looking at neurons and glial cells in the brain, so this project aims were to create a teaching tool that combined the cell characteristics from numerous sources and unify the study of several key types of cranial cells. Using and manipulating 3D bracketed L-Systems for each cell enabled the display of unique characteristics that could be easily compared in a 3D environment. To solidify the user’s knowledge of these cells, a brain atlas was added to show their location at two major cross-sections, images of real and illustrated cells were inserted for comparison, and an information box provided supplementary information. The end result successfully visualized three types of neurons and three types of glial cells in a way that resembled their real counterparts, showed varying levels of detail, and enabled production of diverse but still morphologically similar cells. This provides a good foundation for future expansions on the teaching tool, including the option to include more cells and modify their production rules as more information becomes known and accepted. 1. INTRODUCTION AND MOTIVATION Neuroanatomy is a vast, complicated area of study that is important for various topics in biomedical research. Unfortunately, finding consistent information is often times difficult due to an abundance of obsolete findings mixed in with new results. Even the main divisions of the brain seen in hundreds of online search results and textbooks have inconsistencies that experts disagree with. This lack of consistency in visual and numerical data motivated me to narrow down on a topic that shows significant conflict, and create a teaching tool that will focus on providing relevant and accurate (in terms of most recent knowledge) information without the need to search through numerous dubious resources. The focus of my project is cells in the human brain. Out of several possible topics, the characteristics of neurons and glial (support) cells found in the brain were most troubling. Since students often use online resources for their studies, especially since textbooks are often outdated in this field, it was alarming to see research completed over five years ago on proportions of neurons to glial cells greatly overshadowed by older notions of the human brain’s cellular proportions [1]. Tatiana Karaman Figure 1. Three different artistic representations of Oligodendrocytes [2][3][4] Figure 1 shows a more relevant example; an online search for Oligodendrocytes was conducted, and all three images were found on the very first page of results. Although some similarities are present, the problem arises when one needs to learn about other, structurally similar but not identical cells. Often times, a resource provides information on only one or two cell types, and if three different sources show such differences for one type, one can see how varied the information for several types of cells from different sources can get. This project proposes a way to unify the study of cranial cells by researching the distinctive characteristics of several prominent neuronal and glial cells of the human brain, and using Lsystem techniques to visualize these differences for an easy comparison of visual and numerical information within an educational tool. 2. BACKGROUND RESEARCH The background research for this project consists of two distinct components: past use of Lsystems for biological visualizations, and relevant characteristics found for the cells that were chosen for this teaching tool. 2.1 L-Systems in Biological Visualizations L-Systems have been seen in a number of biological visualizations, most of which concentrated on plants such as trees, bushes and flowers, and the scenes and images that can result from their combinations [5][6]. There have also been several projects on modeling the growth of neurons using L-Systems and growth cones, with a 2D example shown in Figure 2. Tatiana Karaman Figure 2. Growth of Purkinje cells [7] The project that seemed closest to the goals I was trying to achieve is L-Neuron, a tool that generates neuronal analogs by stochastically sampling parameter values from experimental distributions [8]. In one example, the tool achieved a very close approximation to pyramidal cells based on the experimental distributions taken from a 1979 paper. However, as indicated by the comparison between a generated and real Purkinje cells in Figure 3, one can see that either the values in the table are outdated, or that the biological variety of cells is at times hindered by rigid constraints in algorithms. This report will try and demonstrate that altering the basic, differentiating characteristics between cells can be enough to provide an outcome that successfully teaches neuronal and glial cell structures. Figure 3. Left: Purkinje Cell modeled using L-Neuron Right: Real Purkinje Cell [9] 2.2 Cell Characteristics There are dozens of recognizably unique cells in the human brain, and from hundreds to thousands more that we are not completely aware of or know much about yet. For this project, I chose to concentrate on three types of neurons and three types of glial cells that are Tatiana Karaman prominent in research and education. Table 1 shows an assortment of information I found most important to consider when visualizing these cells. Some approximate cell sizes were taken from scale images in cases where no credible data could be found. Cell Type Granule Size ~30µm Pyramidal Microglia ~100-200µm in width, varied length ~200µm1mm [13] Can range ~500-700µm in length or more [15] ~30µm [17] Oligodendrocyte <25µm <10µm Astrocyte ~25µm [20] ~10µm [20] Purkinje Soma ~10µm [10] ~20µm Triangular soma Dendrites 3-4 Claw-like endings[11] 1 long apical dendrite and several shorter, heavily branched basal dendrites, all covered in spines [14] Axon Y ~80µm Usually one very heavily branched dendrite that forms a nearly 2D layer [16] Elongated and elaborate in gray matter, more compact in white matter [18] Slender, less branched than Astrocytes [19] 5-10 branches, evenly distributed[20] Y Purkinje layer of cerebellum [11] N Throughout the CNS, more in gray matter than white matter [18] N White matter [11] N Protoplasmic: gray matter Fibrous: white matter Radial: between gray matter and pia mater [11] ~10µm [17] Y Location Granule cell layer of cerebellum [12] Forebrain structures [13] Table 1. Important cell characteristics 3. IMPLEMENTATION DETAILS 3.1 Choice of L-System The L-System I decided to implement is a bracketed, context-free 3D system with a randomized turtle interpretation. A 3D representation was a clear choice, as 3D visualizations of biological systems are much more realistic, and allow us to view the differences in the organization of the structure from many angles (Purkinje cells, for example, are much flatter than many other neurons). A bracketed system allows for branching, and since brackets within brackets are also possible, this can show obvious comparative differences between lightly and heavily branched structures. Finally, I chose to randomize turtle interpretation instead of randomizing the LSystem itself. Stochastic L-Systems might be a future direction, but they are not always needed, as some cells are best represented with a specific set of unchanging rules (such as the claw-like dendrites on granule cells. For now, all visualized cells vary the angles of the turtle’s heading, some will vary the number of branches created (by altering the number of active turtles), and some will show variety in branch length. Tatiana Karaman 3.2 L-System Vocabulary Based on turtle implementations found in the Algorithmic Beauty of Plants [21], we can extend the state of a standard 2D turtle from a triplet (x,y,α) to a 3D state with variables (x,y,z,α,δ,φ), where (x,y,z) are 3D coordinates for the position of the turtle, and (α,δ,φ) are three angles that rotate the heading of the turtle on each axis. With minor modifications, the symbols used to denote all components of the cells, the relevant rotational matrices, and order of rotation are listed below: F/L/S = Move forward by a set distance, draw line or cylinder G = Extends main branch B = Draws bottom cone for cell body to axon/dendrite connection T = Draw terminals/synapses at current point K = Extend line/cylinder upwards, branch twice, draw synapses on ends [ = Push current state of turtle onto a stack ] = Pop saved state from stack and make current state + = Turn left by α, use rotation matrix RU(α) - = Turn right by α, use rotation matrix RU(-α) & = Pitch down by α, use rotation matrix RL(δ) ^ = Pitch up by α, use rotation matrix RL(-δ) < = Roll left by α, use rotation matrix RH(φ) > = Roll right by α, use rotation matrix RH(-φ) Order of Rotations: RH -> RL -> RU Rotation Matrices: RU(α) = cos(α) sin(α) -sin(α) cos(α) 0 0 RL(δ) = cos(δ) 0 sin(δ) RH(φ) = 1 0 0 0 1 0 0 0 1 -sin(δ) 0 cos(δ) 0 0 cos(φ) -sin(φ) sin(φ) cos(φ) 3.3 System and Notable Classes This teaching tool application was written in C++ using OpenGL and freeglut. Tatiana Karaman InfoBox Class This class was created for the efficient addition of information about a cell in a way that creates a translucent overlay but still allows the text and the cell to be seen at the same time. Turtle Class This class is for initializing the new turtle. It requires a 3D point and three angles that are used for heading calculations. Turtle stores these variables, and modifies the angles each time a new angle is specified for any of the three directions. Additionally, it uses the point and angles and inputted distance to calculate a new location of the turtle whenever the forward() function is called. TurtleGraphics Class This class draws all the necessary objects based on the position and heading information it receives from Turtle. It stores the color and branch length, saves the state of the turtle onto a stack in bracketed operations, sends updated angle information to Turtle, and contains functions for low resolution and high resolution dendrites, axons, terminals and other parts of the cell which get called depending on the distance of the camera from the cell, and the type of cell being drawn. Neurons - main OpenGL render loop All axioms and production rules for each cell are specified here, as well as functions for drawing the cell bodies. The production rules have several options they can call based on the distance the user is from the center of the cell. The main render loop is also responsible for the creation and navigation of both windows, and the generation of vectors containing the necessary random values that allow for variation in the modeled cells. 3.4 Formation of Cell Parts Looking at the cell characteristics listed in the Background Research section, information such as number and qualities of branches, shape of cell body, size and overall shape of the cell and prominent components should all be taken into account when creating the cells. These numbers will be most important in dictating the relative proportions, number of turtles running for each cell, and the range for rotational angles. 3.4.1 Cell Body Most neurons and glial cells have a cell body (soma) that is spherical, or close to spherical in nature, with wider, cone-like extensions in areas where dendrites or axons protrude. Tatiana Karaman Figure 4. From left to right: Astrocyte [22], Purkinje cell body [23], Pyramidal cell [24] Figure 4 shows two examples of these cell bodies (Astrocyte and Purkinje cell), as well as a pyramidal cell that is known by its distinguishable soma in the shape of a rounded pyramid. For the teaching tool, the pyramidal cell body was created using the sphere and cone primitives, combined together to match the shape seen in literature. The cell bodies for all other cells were using spheres or ovals, with additional cones drawn at the connection point between every soma and its axons and dendrites (these cones are specified in the L-System vocabulary as ‘B’). Examples of both types of cell bodies as seen in my tool are shown in Figure 5. As the user zooms closer to the center of the cell the polygon count of the shapes increases, providing a smoother result. Figure 5. Left: Pyramidal cell body, right: Microglial cell body 3.4.2 Axon and Dendrites Most neurons have a single axon, and all three kinds represented by this project have these long, slender and unbranched projections. Table 2 lists all of the axioms, production rules and other important variables for the axons and dendrites of all six types of cells. All systems containing the letter ‘L’ create axons, while all other systems create dendrites. Tatiana Karaman Cell Name Cell Type n Axiom Production Rules # of Turtles α, δ, φ ranges Granule Cell Neuron 2 #1: BF F = F<^-FX X = [<^-XT][>^-XT][<^-XT] 3-4 [-30,30] [-30,30] Pyramidal Cell Neuron 2 #2: BL #1: BF L=L+L-L F = FF[<^-FT]F[>&+FT[>&+FT]] 1 3-5 [2,10] [-40,40] #2: G G = G<^+G[<^+GT]G[<^+GT] 1 4 #3: BL #1: BF L=L+L-L F = F[+^SK][^-SK] S = F[+^SK][^-SK] 1 1 [-2,2] [-40,40] [2,10] [5,25] [35,55] 2 2 2 #2: BL BF BF BF L=L+L-L F = F[<^-FT]^F[>&+FT] F = F[<^-FT] F = F[<^-FT][>&+FT] 1 8-12 7-12 5-10 Purkinje Cell Microglia Oligodendrocyte Astrocyte Neuron Glial Glial Glial [2,10] [-40,40] [-40,40] [-40,40] Table 2. L-System axioms, production rules, angles and other important variables for creation of axons and dendrites for select neuronal and glial cells. Consult section 3.2 for symbol definitions. The Production rules seen in Table 2 were meant to mimic characteristics discussed in previous sections. For example, the production rules for ‘X’ in granule cells allow the dendritic endings to always appear claw-like. On the other hand, the production rules for Astrocytes provide the desired evenly distributed branching, while the rules for Oligodendrocytes show a less branched and more slender result (Figure 6). Figure 6. Branching comparison of Oligodendrocytes (left) and Astrocytes (right) Pyramidal cells are also an interesting case; they are composed of apical and basal dendrites. While the 3-5 basal dendrites show heavy, instant branching that can be easily created using a number of bracketed production rules, the apical dendrite needs to be long and gradually branching as the dendrite grows further away from the cell body. Additionally, pyramidal cell dendrites are covered in dendritic spines seen in Figure 7, which were also visualized in my tool using simple spheres. Tatiana Karaman Figure 7. Left: Illustrated example of dendritic spines and a stained dendrite [25] Right: Dendritic spines as seen in my teaching tool Purkinje cells are quite different from the rest, as they are flat, almost 2-dimensional in shape, and fill up a very large amount of space until they reach the end of the Purkinje layer in the cerebellum. The chosen method of representation was to begin with production rules that quickly and widely branch out the dendrites, then have a function that draws an additional, long branch with two sub-branches upwards to a presumed end of the Purkinje layer. The result, shown in Figure 8 is a mostly flat, space filling dendritic forest. Figure 8. Dendrites of a Purkinje cell Tatiana Karaman 3.4.4 Axon Terminals and Dendritic Synapses Axon terminals and dendritic endings should be seen at the end of every axon and dendritic branch, respectively. Figure 9. Interaction between an axon terminal and a dendritic ending [26] Figure 9 shows how these terminals and endings typically look; their goal is to communicate with neurotransmitters and relay electrical signals from cell to cell. In my visualization, these are represented by simple points in lower-detail distances, and by spheres as we get closer. Figure 10 demonstrates this change. Figure 10. Change from lower-detail to higher-detail representation 4. USER INTERFACE The user interface is very straightforward and easy to use. It consists of two windows (as seen in Figure 11); the first window is the “Brain Atlas” and the second window is the “Cranial Cell Modeler”, the latter of which is what the user interacts with. Tatiana Karaman Figure 11. User Interface The Brain Atlas highlights the location of the selected cell in the saggital and coronal crosssections. In some cases, highlights with different alpha values represent low and high density locations of the cell. In this version of the tool, the brain images were taken from online resources and individually modified to reflect current knowledge of cell locations [27][28]. There are also two images below the brains that help connect the 3D representation to an image of a real cell, and an illustration that is commonly found in textbooks or online sources. The Cranial Cell Modeler has a number of possible interactions listed below. - Choose cell: use the white arrows on the bottom left of the window o For different Astrocyte types, click on one of the three pop-up buttons See a different representation of the same type of cell: press R Get more information: select white box on the bottom right of window Rotate around cell: hold left mouse button and move mouse Zoom towards/away from cell: hold middle mouse button and move mouse up/down Tatiana Karaman 5. RESULTS The teaching tool is able to visualize three different types of neurons, and three different types of glial cells, examples of which are shown in Figures 12-17. Figure 12. Granule Cell Tatiana Karaman Figure 13. Pyramidal Cell Figure 14. Purkinje Cell Tatiana Karaman Figure 15. Microglial Cell Figure 16. Oligodendrocyte Tatiana Karaman Figure 17. Protoplasmic, fibrous and radial Astrocytes Additionally, the angle values from Table 2 were used to produce random sets of parameters within those ranges, allowing each redraw to take from the regenerated parameter sets and create a variety of cells that remained structurally similar. Figure 18 successfully demonstrates this for granule cells. Figure 18. Variations of a granule cell Tatiana Karaman 6. FUTURE DIRECTIONS There are several directions this teaching tool can be taken to improve its accuracy and visual appeal. Looking at leading research in the field would allow us to improve on the current algorithms and yield more accurate results, and agreement between researches and what is currently taught in classrooms would also narrow down the chance of serious errors and misrepresentations in this tool. Some other possible work includes the addition of more cells to the tool, the presence of more information on each cell and/or links to outside resources, labels on important components of the cell and within the brain atlas, and more aesthetically pleasing visualizations. These can be achieved by replacing lines and cylinders with curves, and creating smoother shapes by merging the round cell bodies with the cones that represent dendritic protrusions. Even further down the line, the tool could greatly benefit from features such as interactions between cells, showing the cells at different states (such as activated and inactivated microglia), and showing cell deterioration or alteration due to ailments or diseases. 7. CONCLUSION The current version of this teaching tool allows the user to learn about three different types of neurons and three different types of glial cells in the human brain in a three-dimensional space. The user is able to see the approximate scale of each cell, compare the differences between the different types of cells and appreciate the similarities of each re-created cell of the same type, and see the cells with various levels of detail. Additionally, the user is able to learn more information about each cell including its characteristics and locations in the brain, and finally compare the results to examples of real and illustrated cells. There is still much room for improvement and numerous possibilities for future work, but for now the goals of this project were successfully accomplished and my modification of L-Systems proved to be an effective method for visualizing several kinds of neurons and glial cells of the human brain. Tatiana Karaman 8. REFERENCES [1] Azavedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009; 513(5):532-41 [2] EurekAlert [homepage on the Internet]. AAAS [updated 2013; cited 2013 Dec 12] Available from: http://media.eurekalert.org/multimedia_prod/pub/web/34778_web.jpg [3] Motifolio [homepage on the Internet]. Motifiolio Inc. [updated 2013; cited 2013 Dec 12] Neuroglial cells Oligodendrocyte. Available from: http://site.motifolio.com/images/Neuroglialcells-Oligodendrocyte-7111140.png [4] Journal of Cell Biology [homepage on the Internet]. The Rockefeller University Press [updated 2013; cited 2013 Dec 12] Available from: http://jcb.rupress.org/content/164/1/111/F8.large.jpg [5] Streit L, Federl P, Sousa MC. Modeling Plant Variation through growth. Comp Graph Forum. 2005;24(3):497-506 [6] Palubicki W, Horel K, Longay S, Runions A, Lane B, Mech R et al. Self-organizing tree models for image synthesis. ACM Transactions on Graphics. 2009;28(3),58:1-10 [7] Hamilton P. A language to describe the growth of neurites. Biol Cybern. 1993;68:559-565 [8] Ascoli GA, Krichmar JL. L-neuron a modeling tool for the efficient generation and parsimonius description of dendritic morphology. Neurocomputing. 2000;32-33:1003-1011 [9] Le TD, Shirai Y, Okamoto T, Tatsukawa T, Nagao S, Shimizu T, Ito M. Lipid signaling in cytosolic phospholipase A2α–cyclooxygenase-2 cascade mediates cerebellar long-term depression and motor learning. Proceedings of the National Academy of Sciences. 2010;107:3198-3203 [10] Hirano T, Kubo Y, Wu MM. Cerebellar granule cells in culture: Monosynaptic connections with Purkinje cells and ionic currents. Neurobiology. 1986;83:4957-4961 [11] Diamond MC, Scheibel AB. The Human Brain Coloring Book. 19th ed. HarperCollins Publishers. 1985. [12] Contestabile A. Cerebellar granule cells as a model to study mechanisms of neuronal apoptosis or survival in vivo and in vitro. Cerebellum. 2002;1(1):41-55 [13] Scholarpedia [homepage on the Internet]. Nelson Spruston [updated 2009; cited 2013 Dec 12] Pyramidal neuron. Available from: http://www.scholarpedia.org/article/Pyramidal_neuron Tatiana Karaman [14] Romand S, Wang Y, Toledo-Rodriguez M, Markram H. Morphological development of thicktufted layer-IV pyramidal cells in the rat somatosensory cortex. [15]Fatemi SH, Halt AR, Realm G. Purkinje Cell Size is Reduced in Cerebellum of Patients With Autism. Cell Mol Neurobiol. 2002. 22(2):171-175 [16] Kaneko M, Yamaguchi K, Eiraku M, Sato M, Takata N, Kiyohara Y et al. Remodeling of monoplanar Purkinje cell dendrites during cerebellar circuit formation. PLoS One. 2011;6(5):113 [17] Weimer RM, Lozlowski C. An automated method to quantify microglia morphology and application to moitor activation state longitudinally in vivo. PLoS One. 2012;7(2):1-9 [18] Tremblay ME, Stevens B, Serra A, Wake H, Bessis A, Nimmerjahn A. The role of microglia in the healthy brain. J Neurosci. 2011;31(45):16064-16069 [19] Pannesse E. Neurocytology: fine structure of neurons nerve cell processes, neuroglial cell. 1st ed. Thieme;1994 [20] Sofroniev MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119(1):7-35 [21] Prusinkiewicz P. The algorithmic beauty of plants. 1st ed. Springer-Verlag:1996 [22] Neuroscience Gallery [homepage on the Internet] Conncad c2010-2013 [updated 2013; cited 2013 Dec 12] Veterinary Neurohistology Atlas Catalog, Astrocytes (Golgi stain). Available from: http://www.conncad.com/gallery/single_cells.html [23] Neuroscience Gallery [homepage on the Internet] Conncad c2010-2013 [updated 2013; cited 2013 Dec 12] Synaptic connection between an interneuron and a postsynaptic Purkinje cell in the cerebellar cortex. Available from: http://www.conncad.com/gallery/single_cells.html [24] Carey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM et al. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry. 1998;65:446-453. [25] Neuroscience Gallery [homepage on the Internet] Conncad c2010-2013 [updated 2013; cited 2013 Dec 12] ] Dendritic Spines and Microtubules. Available from: http://www.conncad.com/gallery/spines_synapses.html [26] Scientific American [homepage on the Internet]. Scientific American, a Division of Nature America, Inc [updated 2013; cited 2013 Dec 12] Available from: http://www.scientificamerican.com/article.cfm?id=keeping-up-with-the-neurons Tatiana Karaman [27] Visual Photos [homepage on the Internet] [cited 2013 Dec 12] saggital section through a human brain. Available from: http://www.visualphotos.com/image/1x6009440/sagittal_section_through_a_human_brain [28] Anatomy of the Brain [homepage on the Internet] Claudia Krebs - University of British Columbia c2006-2012 [updated 2012; cited 2013 Dec 12]. Coronal Brain Sections. Available from: http://www.neuroanatomy.ca/cross_sections/sections_coronal.html