Gene regulatory network of auxin signaling
advertisement

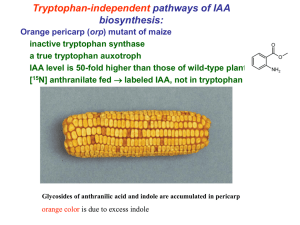
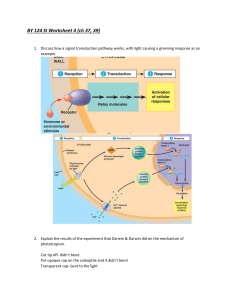
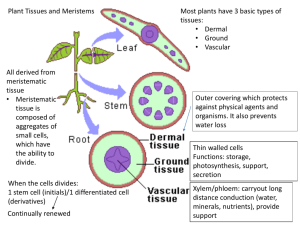
Transcriptional networks controlling hormones responses in Arabidopsis thaliana Plants are built from numerous different types of cells, each performing specific functions to support growth, development, survival and reproduction. Little is known how cells are programmed and different cell types specified. The phytohormone auxin plays a critical role in regulation of cell division and differentiation during plant development through transcriptional regulation of specific genes. It is of fundamental relevance to determine how transcription factor networks are set up and function to enable hormone response cascades and subsequent physiological and developmental responses. Key regulators of auxin-mediated gene expression are 22 ARF and 29 Aux/IAA proteins. The combinatorial action of these factors determines the different characteristic developmental responses. Recent investigations using microarray approaches demonstrated that not all Aux/IAA genes are regulated by auxin and that other factors (besides auxin) participate in their regulation. Moreover, auxin regulates not only Aux/IAA genes but also other transcriptional factors. Here we plan to elucidate regulatory networks by applying chemical genetics using single Arabidopsis cells. Time series analysis of the transcriptome of these cells allowed to define the topology of regulatory networks controlling hormone action. We managed to define the topology of gene networks acting in auxin signaling. 52 transcription factors, differentially regulated by auxin, were found including homeobox genes and members of other families. In this project we will, by combining expression profiling, loss-of-function experiments, chromatinimmunopreciptation (ChIP-chip) experiments, elucidate and model the regulatory network that determines auxin responses. These networks will be extended from single cells to more complex organs to model and simulate auxin induced programs in plant development.