5.5 Control Mechanisms copy
advertisement
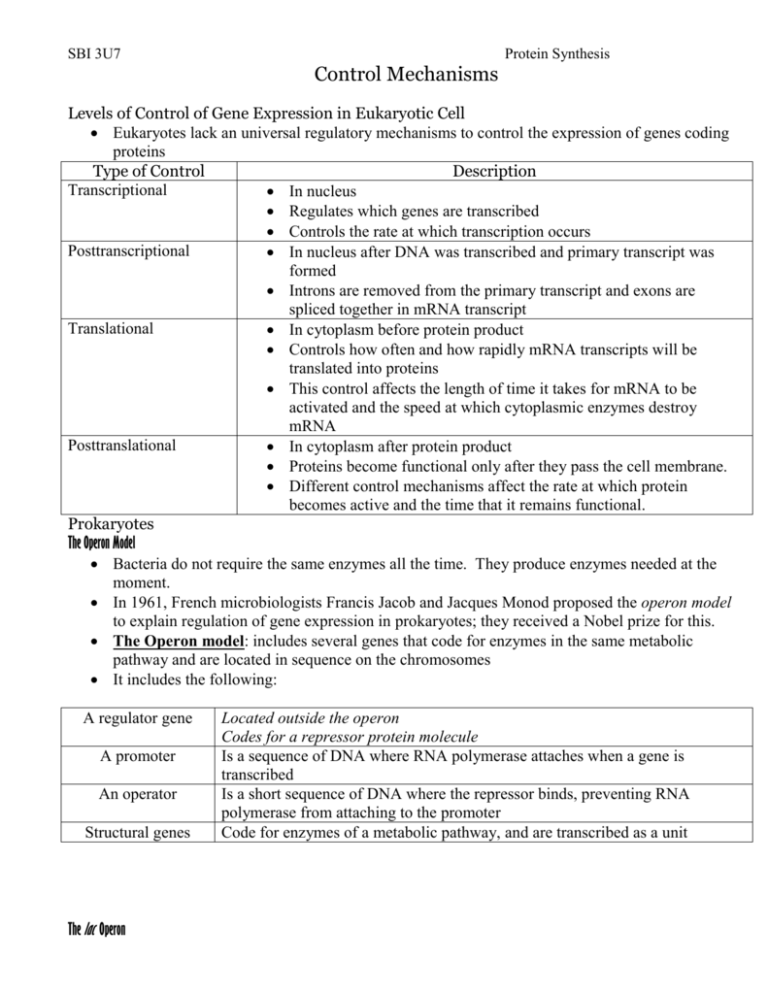
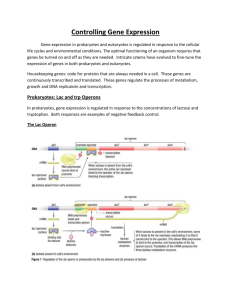
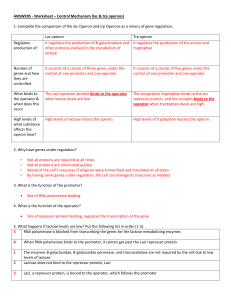
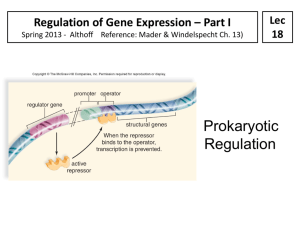
SBI 3U7 Protein Synthesis Control Mechanisms Levels of Control of Gene Expression in Eukaryotic Cell Eukaryotes lack an universal regulatory mechanisms to control the expression of genes coding proteins Type of Control Description Transcriptional In nucleus Regulates which genes are transcribed Controls the rate at which transcription occurs Posttranscriptional In nucleus after DNA was transcribed and primary transcript was formed Introns are removed from the primary transcript and exons are spliced together in mRNA transcript Translational In cytoplasm before protein product Controls how often and how rapidly mRNA transcripts will be translated into proteins This control affects the length of time it takes for mRNA to be activated and the speed at which cytoplasmic enzymes destroy mRNA Posttranslational In cytoplasm after protein product Proteins become functional only after they pass the cell membrane. Different control mechanisms affect the rate at which protein becomes active and the time that it remains functional. Prokaryotes The Operon Model Bacteria do not require the same enzymes all the time. They produce enzymes needed at the moment. In 1961, French microbiologists Francis Jacob and Jacques Monod proposed the operon model to explain regulation of gene expression in prokaryotes; they received a Nobel prize for this. The Operon model: includes several genes that code for enzymes in the same metabolic pathway and are located in sequence on the chromosomes It includes the following: A regulator gene A promoter An operator Structural genes The lac Operon Located outside the operon Codes for a repressor protein molecule Is a sequence of DNA where RNA polymerase attaches when a gene is transcribed Is a short sequence of DNA where the repressor binds, preventing RNA polymerase from attaching to the promoter Code for enzymes of a metabolic pathway, and are transcribed as a unit SBI 3U7 Protein Synthesis Lactose Disaccharide found in milk Consists of two sugars: glucose and galactose E.coli bacterial cells found in the intestinal lining can use the energy supplied by lactose for growth. To be able to use this energy E.coli must split lactose into glucose and galactose. E. coli produces an enzyme called -galactosidase o By using a negative regulation control the transcription and translation of the -galactosidase gene (in the bacterial cell) produce the enzyme only when necessary. Regulatory Gene Promoter Operon Structural genes lacZ mRNA protein Encodes the enzyme -galactosidase lacY Encodes the enzyme -galactosidase permease lacA Encodes the enzyme transacetylase Repressor -blocks/activates the transcription of the -galactosidase gene by binding/no binding to the lactose operator (signal molecule or inducer) Lactose is present Repressor Protein Lactose is absent lactose glucose + galactose facilitates entry of lactose in cell accessory in lactose metabolism SBI 3U7 Protein Synthesis Repressor Protein The trp Operon Jacob and Monod found some operons in E. coli that exist in the “on” condition rather than the “off” condition. E.Coli bacteria use the amino acid tryptophan to produce protein. This prokaryotic cell produces five polypeptides to yield three enzymes to synthesize the amino acid tryptophan. In the trp operon, the regulator (upstream) codes for a repressor protein that has a binding site to tryptophan When tryptophan is in abundance, it will bind to the repressor protein allowing the repressor to bind to the operator. This will inhibit transcription. Since you need trp to activate the repressor, Tryptophan is called a Corepressor. Tryptophan is absent Homework: p. 258 #1-6