File
advertisement

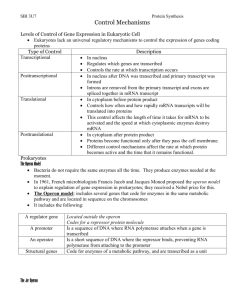
Controlling Gene Expression Gene expression in prokaryotes and eukaryotes is regulated in response to the cellular life cycles and environmental conditions. The optimal functioning of an organism requires that genes be turned on and off as they are needed. Intricate sstems have evolved to fine-tune the expression of genes in both prokaryotes and eukaryotes. Housekeeping genes: code for proteins that are always needed in a cell. These genes are continuously transcribed and translated. These genes regulate the processes of metabolism, growth and DNA replicaiotn and transcription. Prokaryotes: Lac and trp Operons In prokaryotes, gene expression is regulated in response to the concentrations of lactose and tryptophan. Both responses are examples of negative feedback control. The Lac Operon The Lac operon is a cluster of three genes that code for the proteins involved in the metabolism of lactose. There are three key regions of the operon itself (promoter, operator and genes for metabolizing proteins) and there is a gene for a lac repressor immediately upstream from the lac operon. The gene for the lac repressor (LacI) is continuously transcribed and is always present in the cell. It takes cues from the environment of the cell (concentration of lactose). If lactose is absent, the repressor remains bound to the operator (controls the transcription of the genes for the metabolizing proteins) and transcription is blocked. In the presence of lactose, the lactose binds to the repressor changing its shape and making it inactive. In this state, the repressor can no longer bind to the operator and the metabolizing proteins are made. Lactose itself acts as a signal molecule telling the cell when to synthesize the lactose metabolizing proteins. This makes lactose an inducer because it serves to initiate the production of the enzymes. The trp Operon Tryptophan is an important amino acid that prokaryotes can either make or take up from the environment. The trp operon tells the cell if it has to make its own tryptophan. The trp operon has the same structure of the lac operon but the activity of the repressor is different. In the lac operon the presence of the lactose caused the repressor to be released, in the trp operon, the presence of tryptophan causes the repressor to bind to the operon. When tryptophan is absent, the repressor gene is in the inactive state and does not bind to the operator and tryptophan is synthesized by the cell. When tryptophan is present, it binds to the repressor and changes the shape so that it can bind to the operator region of the operon. In the trp operon, tryptophan acts as a corepressor because binding of tryptophan to the repressor allows the repressor to bind to the operator. Eukaryotes Gene expression in eukaryotes is more complex because protein synthesis occurs in more than one step. Eukaryotes do not use the operon system as prokaryotes do, rather there are four different categories of gene expression mechanisms in eukaryotes. Transcriptional regulation DNA organization: because the DNA is wrapped around histones, not all the promoter regions are accessible for the proteins that initiate transcription. Inorder to unwrap from the histones, activator molecules may bind to the DNA exposing the promoter region, or repressor molecules may bind to the DNA to conceal the promoter or to stop transcription. Methylation (a methyl group is added to the cytosine bases in the promoter region of a gene inhibiting transcription) through a process called silencing Post-transcriptional Regulation: Alternative Splicing: produces different mRNAs from pre mRNA by removing different combinations of introns. For each member of a protein family, introns and exons are different. This method is used to optimize the production of different proteins depending on the cell type. Masking proteins: keep mRNAs in an inactive form until it is time for that mRNA to be translated. This is common in animal eggs that have the mRNA ready in case of fertilization. Hormones are a regulatory molecule that will directly or indirectly affect the rate of degradation of a particular mRNA… Translational Regulation: Changing the length of the poly-A tail: adding or removing extra A bases may increase or decrease the time required for the translation of the protein. Post-Translational Regulation: Processing: proteins exist in the cell in precursor forms and processing mechanisms remove particular sections of the protein and activate it. Ex: Preinsulin Chemical modification: certain chemical groups are added to or removed from a protein affecting its function. The presence or absence of the chemical groups puts the protein on hold until it is needed. Degradation: proteins with short life spans are marked with ubiquitin. This is recognized by the degradation mechanisms of the cell. By adding and removing these markers, the lifespan of the cell can be either shortened or extended.