File
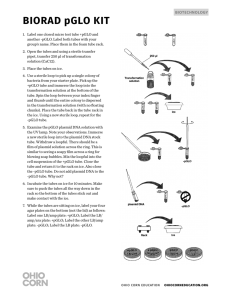
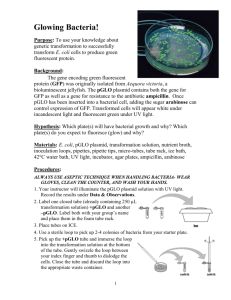
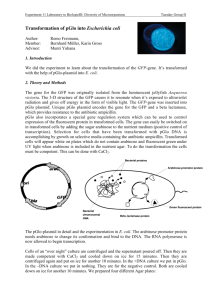
advertisement

Stephanie Bohr pGlo Bacterial Transformation Lab Period A Introduction The purpose of this experiment is to see how bacterial cells be genetically transformed with plasmid DNA containing a jellyfish gene. If the bacteria contain arabinose it will make green fluorescent protein (GFP), then it will glow. The independent variable is the GFP and the dependent variable is whether or not it will glow. Plasmid occurs naturally in many bacteria. Plasmid is small circular DNA molecules. They contain few genes are much smaller than the bacterial chromosomes which are highly compacted structures and share many properties with their eukaryote counterparts. Plasmid DNA is replicated inside bacterial cells. You will also be using pGLO which is a recombinant plasmid. It has been engineered with many different genes, including one form a bioluminescent (glowing) jellyfish. There is also GFP or known as green fluorescent protein. It is a protein that glows a green color when exposed to ultraviolet lights. It is seen only when in the presence of sugar arabinose which is a sugar that activates the production of GFP. Materials UV Lamp Two micro centrifuge tubes Foam tube rack Marker Calcium chloride solution 5 sterile transfer pipettes Plastic cup for biohazard waste containing 10% bleach solution Crushed ice 2 foam cups Starter agar plate with colonies of E. coli 6 sterile inoculating loops pGLO plasmid DNA hot and cold tap water thermometer LB nutrient broth 4 agar plates Clock or watch Incubator Procedure 1. Before starting, observe the starter plate (don’t remove the Petri dish lid) with colonies of E. coli. 2. Shine the UV lamp on the colonies and observe them 3. Label one closed micro test tube +pGLO and another –pGLO. 4. Label both tubes with your group’s name. 5. Place them in the foam tube rack. 6. Open the tubes and using a sterile transfer pipette, transfer 250µ of transformation solution (CaCl2) 7. Place the tubes on ice 8. Use a sterile loop to pick up a single colony of bacteria from your starter plate. 9. Pick up the +pGLO tube and immerse the loop into the transformation solution at the bottom of the tube. 10. Spin the loop between your index finger and thumb until the entire colony is dispersed in the transformation solution with no floating chunks. 11. Place the tube back in the rack in the ice. 12. Using a new sterile loop repeat for the –pGLO tube 13. Examine the pGLO plasmid DNA solution with the UV lamp and note observations. 14. Immerse a new sterile loop into the plasmid DNA stock tube. 15. Withdraw a loopful 16. There should be a film of plasmid solution across the ring 17. Mix the loopful into the cell suspension of the +pGLO tube. 18. Close the tube and return it to the rack on ice. 19. Incubate the tubes on ice for 10 minutes 20. Make sure to push the tubes all the way down in the rack so the bottom of the tubes stick out and make contact with the ice 21. When the tubes are sitting on the ice, label the four agar plates 22. Using the foam rack transfer the +pGLO and –pGLO into the hot water bath set at 42 degrees Celsius. 23. Wait 50 seconds and place both tubes back on ice for two minutes 24. Remove the rack containing the tubes from the ice and place on table. 25. Open a tube and using a sterile pipette, add 250µ of LB nutrient broth to the tube and recluse it. 26. Repeat for other tube 27. Let sit at room temperature for ten minutes 28. Tab the closed tubes with your finger to mix 29. Using a new sterile pipette for each tube, add 100µ of the transformation and control suspensions onto the appropriate plates. 30. Use a new sterile loop for each plate and spread evenly around the surface of the agar 31. Stack up your plates and tape them together 32. Store upside down in the 37 degrees Celsius incubator until the next day Next Day 1. Observe the colonies of E. coli cells on the four plates you prepared 2. Observe the E. coli starter plate 3.Use the hand held UV light to look for the presence of the fluorescent protein. Plate Appearance Under UV light LB/amp – No Glow LB – No Glow LB/amp/ara + Glow LB/amp + No Glow Variable held constant: Tubes holding pGLO, bin of cold water, hot bath Observations -pGLO LB/amp No Glow -pGLO LB No Glow +pGLO LB/amp/ara Glowed +pGLO LB/amp No Glow Conclusion In conclusion, we discovered that only bacteria containing arabinose will glow because arabinose is the sugar that activates the production of green fluorescent proteins. The plate that contained the LB/amp/ara was the only one that glowed because it was the only one that contained the arabinose. Our experiment was not valid or reliable because when it came time to spread the pGLO onto the plates we spread it onto the lid not allowing the bacteria to eat off of the stuff inside each plate, this doesn’t allow any bacteria to grow. When we consulted with another group that did it correctly it showed that only the LB/amp/ara glowed where as if we were to look at our results we wouldn’t have been able to see that which would have thrown off all of our results. Other than the problem of the placement of the pGLO, our group followed all of the other instructions properly and was successful. Everything was accurately measured and distributed correctly to each plate, and was held on the ice, heat and room temperature for the correct amount of time. If this experiment were to be repeated and the pGLO was put on the proper side of the plates, the results would come out correctly