Cell_biology_Block_4[1] - CellBioReview
advertisement
![Cell_biology_Block_4[1] - CellBioReview](http://s3.studylib.net/store/data/008534897_1-a83a4c500ff7516bda3d65a2feea2033-768x994.png)
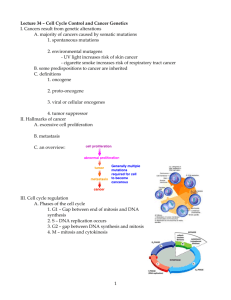
10 from each block Cell biology Block 4 Learn about Cdks, cyclins, CDKI, Wee1 kinase, Cdc25, APC/C and SCF Cdks Cyclin-dependent kinases The progression of the cell cycle through various phases is regulated by transition through specific “check points”, mediated by specific phosphorylating enzymes called CDKs This check point is a type of surveillance system, it allows for detection of: An incomplete previous step Damage to the genome/mitotic spindle When damage is detected, cells arrest at specific phases Allow time to repair the damage Three major regulatory pathways: Start checkpoint: The key checkpoint which determines whether or not the cell will duplicate Regulated by growth factors, nutrients, and integrity of DNA Rb is the primary regulatory protein of this checkpoint Rb binds to E2F (blocks transition) G2/M checkpoint G2 to Mitosis transition is blocked until all the DNA is duplicated Controlled by MPF is blocked until DNA replicated Spindle checkpoint Metaphase to anaphase transition Makes sure that all chromosomes are properly attached to microtubules Anaphase-promoting complex (APC/C) is a key regulatory factor of this checkpoint Growth factors Stimulatory growth factors Activate Ras pathway Ras mutations commonly found in pancreatic, colon, lung, and bladder cancers, and occurs in about 25-30% of all cancers Activate the PI3-kinase-Akt pathway PTEN mutations found in 50% prostate cancer, 35% uterine cancer and also to varying extent in other cancers Inhibitory growth factors act through Cdk inhibitors TGF-beta causes an increase in CDK inhibitor p15 and p21 Downstream effects, increase in production of cyclin D The Cdk inhibitor p21 plays a key role in preventing cells containing damaged DNA from passing through the G1 checkpoint Check points Each CDK phosphorylates and modulates the activity of a subset of target proteins specific for individual transition within the cell cycle Mammalian cells have several CDKs, viz Cdc2 (CDK1), CDK2, CDK3, CDK4, CDK6, and CDK7 Act at different transitions in the cell cycle Cyclins CDK needs to team up with cyclins in order to work And need to be posttranslationally modified correctly by phoshporylation at the right residues Activation of CDKs require their association with another group of proteins called Cyclins Cyclin D,H,E,A,B Contribute to CDK substrate specificity The levels of different cyclins vary during the cycle Ex. Cyclin E accumulates in late G1, associates with CDK2 and is destroyed as cells enter S phase The activity of cyclin-CDK complex is further subject to positive or negative regulation Phosphorylation/dephosphorylation Inhibitors of CDKs Proteolysis of cyclins and inhibitors APC/C Cyclin B levels rise just before mitosis APC/C marks cyclin B with a ubiquitin for it to be degraded by proteasomes The regulation of CDK activity by inhibitory phosphorylation The active cyclin-cdk complex is turned off when the kinase wee 1 phosphorylated two closely spaced sites above the active site Removal of these phosphates by the phosphatase cdc25 activates the cyclin-cdk complex Cdk activating kinase adds the activating phosphate – – – – – CDK1 The cyclin D/CDK4/CDK6, cyclin E/CDK2, and cyclin A/CDK2 are inhibited by a group of CDK inhibitor proteins (CDK1) that include p21 and p27 CDK1s also impair CDK activating kinase activity CAK APC/C and SCF Anaphase promoting complex or cyclosome (APC/C) is a ubiquitin-ligase that marks the S and M cyclins for destruction APC/C activity changes depending on whether it is interacting with the subunit Cdc20 during anaphase of Cdh1 from late mitosis into G1 APC/C is off most of the cell cycle because it has to team up with CDC20, which is usually off as well Following its activation in mid-mitosis, the APC/C remains active in G1 When the G1/S-Cdks are active in late G1, APC/C is turned off SCF activity is constant during the cell cycle Ubiquitylation by SCF is controlled by changes in the phosphorylation status of its target proteins Requires the presence of the F-box protein Cdc25 phosphatase Removes inhibitory phosphates from Cdks three family members Cdc25 A, B, and C in mammals primarily involved in controlling cdk1 activation at the onset of mitosis Wee 1 kinase Phosphorylates inhibitory sites in cdks Primarily involved in suppressing Cdk1 activity before mitosis P21 Suppresses G1/S-cdk and s-cdk activities following DNA damage Learn in detail the three key regulatory pathways of the cell cycle – G1/S Know the roles of Rb, Cdk4/Cyclin D, Cdk6/Cyclin D and Cdk2/cyclin E, p21, CAK, E2F, DP1 Restriction point control: G1 to S progression The transition from G1 to S phase is regulated by a checkpoint called “restriction point” or start A very important regulatory step, to ensure repair of genome damage before initiation of DNA replication. It is now committed to replicating itself! The initiation of cycle, ex: entry of cells into G1 phase, is determined by extra-cellular signals (mutagens, nutrients, and growth factors) Growth factors induce synthesis of D-type cyclins (D1, D2, D3) D cyclins associated with CDK4 and CDK6 in G1 Cyclin D/CDK4/CDK6 complexes are activated through phosphorylation by an enzyme complex called CAK (CDK activating kinase) Often upregulation of Cyclin D Rb controls the G1 to S transition The retinoblastoma protein, Rb, is phosphorylated by Cyclin D/CDK4/CDK6, which is necessary to drive the cell past the restriction point CDK4 Cyclin e/CDC2 and CDK6 with cyclin D phosphorylates Rb and turns it off Once the cell crosses the restriction point, mitogenic stimulation is no longer needed, and the entry into the S phase is ensured Rb is a tumor suppressor, so it halts proliferation and cell growth Phosphorylation of Rb dissociates E2F, which then heterodimerizes with DP family of transcription factors When Rb binds E2F, it turns off transcription Rb is active when hypophosphorylated, and turned off when hyperphosphorylated The e2f-DP heterodimeric transcription factors bind to sequences in the regulatory regions of genes important in the control of cell growth Ex: c-myc, DHFR, c-myb, cdk1, E2F-1, Cyclin A The synthesis of E2F is upregulated in late G1, presumably through E2F. Restriction point control and S phase Cyclin E/CDK2 kinase is needed to maintain Rb in its hyperphosphorylated state As cyclin E/CDK2 activity decreases, cyclin A synthesis is induced Accumulation of the cyclin A/CDK2 complexes signals entry into S phase In HPV There are 2 oncogenes: E6 – compromises p53 E7 – compromises Rb – G2/M Know the roles of MPF (Cdk1/cyclin B) The mitotic cdk1-cyclin B complex (MPF) controls the G2 checkpoint by phosphorylating proteins involved in the early stages of mitosis MPF=maturation-promoting factor CDK1 with cyclin B Activated by multistep process A target: MPF phosphorylates lamin proteins of the nuclear lamina (causing breakup of nuclear membrane) A target: MPF phosphorylates condensin complex which may trigger chromosome condensation Which triggers mitosis, it is the starting point – Spindle assembly check point Know the roles of APC/C, Securin, Separase, Cohesin, Mads, Bubs, Cdc20, MPF (Cdk1/cyclin B) The mitotic Cdk-cyclin complex (MPF) controls the spindle assembly checkpoint by activating the anaphase-promoting complex Controlled by APC/C Targets MPF-cyclin B and securin Anaphase-promoting complex (APC/C) is a ubiquitin ligase that marks proteins for destruction by proteasomes MPF and Cdc20 regulate APC/C APC/C triggers the breakdown of securin Securin normally binds to and inhibits Separin When separin (Separase) is release because of lack of securing, it can target and cleave cohesins, thereby freeing sister chromatids so they can separate from each other Anaphase-promoting complex also triggers the breakdown of MPF cyclin Degradation of cyclin very important for the end of mitosis This process is controlled by proteins associated with the kinetochores Anaphase promoting complex is controlled by kinetochores, which bind Mad and Bub proteins as long as the kinetochores remain unattached to spindle microtubules Mad and Bub proteins inhibits the anaphase-promoting complex by controlling cdc20 activity, thereby preventing the initiation of anaphase Learn about cell cycle arrest in response to DNA damage – Know the roles of p53, ATM/ATR, Mdm2, p21, Myc, Arf – In response to DNA damage, ATM and or ATR trigger the activation of a check point that leads to cell cycle arrest or delay. Checkpoint pathways are characterized by cascades of protein phosphorylation events (indicated with a P) that alter the activity, stability, or localization of the modified proteins. – P53***** (know this pathway)******* A transcription factor Levels kept really low in the nucleus, because a protein called MDM2 binds to it, takes it out of nucleus, brings it to cytosol, marks it with ubiquitin so that it is destroyed P53 plays a pivotal role in cell cycle arrest in response to DNA damage When DNA is damaged, ATM and ATR (2 Dna-dependent protein kinases) leads to the phosphorylation of P53, preventing its degradation Thus, DNA strand breaks by UV or ionizing radiation increase levels of P53 protein P53 is a transcription factor, when it is phorphorylated, the levels increase in the nucleus binding to MDM2 P53 then stimulates the expression of p21 (a CDK inhibitor) that causes cell cycle arrest!! P21 blocks cdk/cyclin complexes LEADS TO CELL CYCLE ARREST So it means the Rb stays in the hyperphosphorylated state It also prevents DNA synthesis by binding to PCNA (proliferating cell nuclear antigen), a subunit of DNA polymerase delta enzyme complex, involved in DNA replication and repair P53 also induces many other genes such as GADD45 and cyclin G that contribute to arrest – Mdm2 Controls the levels of p53 in the nucleus Mdm2 controls the level of p53 in the nucleus by binding p53 and shutting it out of the nucleus where it is destroyed by the ubiquitindependent pathway If p53 is phosphorylated by the ATM/ATR pathway and is acetylated, it will no longer interact with mdm2 Thus, the levels of p53 increase in the nucleus P53 tells the cell to stop, that there is something wrong, either fix it or go kill yourself P53 levels also increase if there is excessive stimulation of mitogenic pathways Mitogen activated pathways lead to upregulation of transcription factor Myc Myc causesArf to be produced Myc is a transcription factor that promotes proliferation Arf inactivates mdm2 No mdm2 results in increased p53 levels Learn about the key differences between Necrosis and Apoptosis (2 types of cell death) Necrosis Swelling and rupturing of injured cells Typically involving many cells at the tissue level as a result of hypoxia, toxins of physical force Necrosis is characterized by severe destruction, cytoplasmic organelle destruction, loss of membrane integrity, heterophagy, accompanying inflammatory response Apoptosis A deliberate genetically controlled cell death (programmed cell death) that occurs in response to specific environmental, developmental, or other stimuli Usually at the single cell level Apoptosis is a neat and tidy process of cell killing, with minimum damage to surrounding cells or tissue, autophagy; without accompanying inflammatory response Why is it important? Ensures homeostasis of all tissues each individual will produce and eradicate a mass of cells equal to its body weight helps prevent tumor growth destroys obsolete or old cells vital control mechanisum in maintaining the immune system death of neutrophils during acute inflammatory response death of B and T lymphocytes after cytokine depletion cell death induced by cytotoxic T cells cell death following certain viral infection (ex. Viral hepatitis) physiological, adaptive, or pathological causes programmed cell death during embryogenesis implantation, organogenesis, involution removal of webbing in between digits, resorption of tadpole tail, pruning of neurons in infant brain development hormone-dependent involution in the adult endometrial breakdown during menstrual cycle regression of lactating breast after weaning cell death following injuries radiation, cytotoxic drugs, hypoxia Stages of apoptosis Morphological characteristics: cell shrinkage, chromatin condensation, surface blebbing, followed by formation of membrane-bound apoptotic bodies containing cytoplasm and tightly packed organelles. Often phagocytosis of apoptotic bodies by adjacent healthy cells and macrophages occurs DNA segregates to nuclear periphery and cytoplasmic volume decreases Cell produces blebs, organelle fragments DNases digest chromatin Cell is dismantled in apoptotic bodies A hallmark of apoptosis: DNA ladder Biochemical characteristics: Protein cross-linking Mediated by transglutaminase Convert cytoplasmic proteins into covalently linked shrunken shells for packaging into apoptotic bodies Protein cleavage mediated by “caspases” Cleavage of nuclear scaffold, cytoskeletal proteins Fragmentation of genomic DNA Mediated by calcium and magnesium dependent endonucleases Triggered by caspases Intracellular zymogens critical for apoptosis Needs to be cleaved in order to be active Genomic DNA is cleaved at internucleosomal sites, into DNA fragments Loss of membrane asymmetry: appearance of phosphotidylserine (a phospholipid), thrombospondin, on outer plasma membrane help recognition of apoptotic cells by macrophages In normal cells, the phosphatidylserines are facing the cytosol During apoptosis, flippases are turned off, so phosphatidylserine is now facing the outside of the cell, so it is just asking to be killed Learn about the role of caspases during apoptosis (Initiator vs effector caspases ) – Caspases Cytosolic aspartate-specific cysteine proteases Caspases are zymogens Need to cleave in order to become active They remain inactive until an apoptosis signal initiates the activation of one (initiator caspase) which will cause a cascade leading to activation of other caspases (effectors) 3 classes of caspases Intiator caspases Caspase 8, 9, 10, and 12 Caspase 8** and 10– death receptor induced apoptosis Caspase 9** – mitochondrial pathway Effector (executioner) caspases Caspase 3, 6, and 7 Inflammatory response mediators Initiators are activated first, and then they go after the effectors Learn about IAPs and anti-IAPs Caspases are regulated by inhibitors (IAPs) The inhibitor of apoptosis proteins (IAPs), XIAP, cIAP-1, c1AP-2 and Survivin, can prevent proteolytic processing of procaspases 3, 6, 7 and 9 Bind to caspases and prevent them from being activated Role of IAPs and anti-IAPs in the control of apoptosis In the absence of an apoptotic stimulus, IAPs prevent accidental apoptosis caused by the spontaneous activation of procaspases The IAPs are located in the cytosol and bind to and inhibit any caspases Some IAPs are also ubiquitin ligases When the intrinsic apoptotic pathway is stimulated, proteins including anti-IAPs are released Anti-IAPs bind and block the inhibitory activity of IAPs are released Now apoptosis can be induced Learn about the 3 classes of BCL-2 family of proteins Bcl2 on outer surface of mitochondria Their job is to tell the cell to stay alive About 30 Bcl2 family members These proteins play tissue-specific as well as signal pathway-specific roles in regulating apoptosis The tissue specificity is overlapping For example, BCL2 is expressed in hair follicles, kidney, small intestines, neurons, and the lymphoid system Whereas Bcl-x is expressed in the nervous system and hematopoetic cells 3 classes: Anti-apoptotic Bcl-2 Bcl-x Bcl-w Proapoptotic – channel-forming Bax Bak Bok Pro-apoptotic BH3 only Bad Bid Bim Roles of Bcl2 family members in apoptosis Bcl-2 which is antiapoptotic, binds bid and blocks formation of channels that allow cytochrome c release from the mitochondria Death signals result in activation of BH3-only protein such as bid, which can lead to mitochondrial pore formation, swelling, and release of cytochrome c Bid binds to and activates the membrane ion-channeled protein Bax, activating cytochrome c release, which binds to Apaf (APF1)and lead to formation of apoptosme Recruits procaspase 9, activate caspase 9, etc. APAF1 is the adaptor this time!!!! Apoptosis triggered by DNA damage** P53 activates/increases production of Bax and PUMA and other factors when the cell has extreme DNA damage Bax and PUMA (death promoting genes) interact with mitochondria Cytochrome C and other factors are released into the cytosol Cytochrome C recruits Apaf1, procaspase 9 and ATP or dATP into a complex (apoptosome) Caspase 9 then stimulates caspase 3 Apoptosis activated Learn in detail two examples of death receptor pathway of apoptosis (TNF and Fas ligand) – Death receptor pathway of apoptosis Activation-induced cell death Has cytotoxic t cells Involves tumor necrosis factor receptor super-family and ligands. Examples: Fas ligand-fas receptor (CD95) TNF-TNFRI TRAIL Pathway: The apoptosis is induced by certain transmembrane or intracellular stimuli Positive induction involving ligands related to TNF binding to their receptors TNF with TNFR1 FasL with FAS/APO-1 Ligand interacts with receptors, the death receptors oligomerizes and recruit adaptor proteins and initiator caspases to a complex, death-inducing complex, which results in activation of initiator caspases. The active initiator caspase will then cleave and activate effector/exevutioner caspases The ligand (either a free ligand or a cell surface-associated protein from another cell) binds to the death receptor, which makes a scaffold for autocatalytic activation of caspases 8 or 10. Active caspases 8 or 10 cleave apoptotic execution caspases directylu However, the pathway also activates Bid, which acts on mitochondrial membrane integrity Apoptosis is induced Cell death receptors binding of ligand to the Fas receptor induces apoptosis by direct activation of caspase-8. Fas ligand consists of 3 polypeptide chains, so its binding induces receptor trimerization. Caspase8 bound to the receptor via adaptor molecules is then activated by autocleavage, leading to activation of downstream caspases and cell death Fas signaling *** (death receptor pathway) Cytotoxic t cell binds to ligand, death receptor ligand Killer lymphocyte interacts with infected cell via CD95/Fas receptors Receptors aggregate and recruit FADD FADD recruits procaspase-8 to complex Interaction inhibited by FLIP FADD is the adaptor protein Procaspase-8 activates each other by cleavage Caspase -8 (initiator caspase) activates caspase-3 via cleavage In some cells, caspase 8 activates Bid, which interacts with mitochondria which induces death Result: Amplify apoptotoic signaling TNF-alpha-TNFR1 induced signals*** (death receptor pathway) TNFR1 can either initiate apoptosis if associated with adaptor proteins TRADD and FADD Stimulate caspase-8 pathway FLIP inhibits apoptosis by blocking the interaction between FADD(it’s in both pathways) and capase-8 and also by activating survival signals Result: amplify apoptotic signaling. Death. -ORInitiate survival response if TRADD is associated with TRAF2 and RIP(no FADD) ** not that the adaptor proteins are different!!!!!! Stimulate NF-kappaB pathway RIP interferes with TRADD interacting with FADD Result: promotes cell survival How these two differ: they differ at the adaptor protein!! Learn in detail two examples of mitochondrial pathway of apoptosis – Mitochondrial pathway of apoptosis (innate pathway) Very important: BCL2 is associated with outer membrane of mitochondria! Key anti-apoptotic protein It’s telling the cell to stay alive Induced by intracellular damage, DNA damage or growth factor withdrawal Release of key mitochondrial proteins into the cytosol from intermembrane space leads to activation of apoptosis A number of stimuli, including chemotherapeutic agents, UV radiation, stress molecules (reactive oxygen and reactive nitrogen species), and growth factor withdrawal appear to mediate apoptosis via the mitochondrial pathway, a death receptor-independent pathway The outer mitochondrial membrane is the target of apoptotic factors Normally cell survival is ensured by balanced expression of survival (=antiapoptotic) and apoptotic factors on the mitochondrial membrane Apoptotic threshold is established, when the level of expression of apoptotic factors (Bak, Bax, Bad, Bcl-xs) exceed the level of expression of survival factors (Bcl2, Bcl-xl, bclw, mcl1) leading to activation of caspases Note: that the survival or apoptotic factors indicated above are members of the bcl2 proto-oncogene family Once the apoptotic threshold is reached, the physiology of the mitochondria changes which leads to the release of calcium and key mitochondrial proteins found in the intermembrane space Cytochrome C, Diablo/Smac, apoptosis inducing factor AIF, endo g, Htra2/Omi – DNA damage and deprivation of survival factor Apoptosis induced by loss of growth factor/survival factor Loss of cytokine binding to receptor initiates second messenger signaling pathway which causes loss of AKT kinase activity Growth factors/hormones are normal survival factors, assumed to suppress preexisting death program Bad and related pro-apoptotic factors are dephosphorylated Bad is a death promoting protein, Phosphorylated bad is usually sequestered in the cytosol bound to 14-3-3 protein, and keeps it away from the mitochondria This occurs when the cell is “wanted” Bad translocates to mitochondria and interacts with bcl2 or bclx Loss of normal mitochondrial physiology Release of cytochrome c and other factors Activation of caspase 9 pathway The 3 ways that extracellular survival factors can inhibit apoptosis – The middle one – what was just discussed above is a MUST KNOW Know the roles of SMAC/Diablo, HtrA2/Omi, AIF and Endonuclease G (Other factors released by mitochondria) FOUND ON outer membrane of mitochondria Calcium Can lead to activation of caspases associated with ER Usually kept very low in the cytosol SMAC/Diablo (anti-IAPs) Directly interacts with IAPs (XIAP, cIAP1 and cIAP2) and block their activity Lead to apoptosis occurring (inhibiting an inhibitor of apoptosis) HtrA2/Omi (anti-IAPs) Blocks IAPs Can cleave IAPs Promotes caspase-independent apoptosis via its serine protease activity Apoptosis inducing factors AIF Transported to nucleus where it stimulates large DNA fragmentation and condensation of chromatin Also involved in electron transport (FAD binding and oxidoreductase activity) Endonuclease G Considered to directly mediate nuclear DNA fragmentation Cytochrome c Learn about cytolytic granule mediated apoptosis Can circumvent the requirement of FAS or TNF receptors Granzymes (A,B,H,K, and M) and porins released by cytotoxic T cells or natural killer cells activate effector caspases inside the cell These enzymes also target other proteins Mcl1 and Bid Leads to activation of mitochondrial apoptotic pathway Lamins Histones Apoptosis and cancer Nearly all have a defect in cell cycle regulation Follicular B-cell lymphoma: chromosomal translocation leads to up-regulation of Bcl-2 Presentation 2- cancer Learn about the role of cell proliferation & differentiation in the genesis of tumors Cell division and differentiation Tumors are produced by uncontrolled cell proliferation in which the balance between cell division and differentiation is disrupted. This is usually coupled with loss of regulated cell death (apoptosis) Cell differentiation is the process by which cells acquire specialized properties that distinguish different cell types from each other As cells acquire traits, they often lose the capacity to divide Normal differentiation process in balanced with cell proliferation so that no net accumulation of dividing cells occur In tumors, this finely balanced arrangement is disrupted and cell division is uncoupled from cell differentiation Consequence: increase in number of proliferating cells which lead to disruption of normal tissue structure and function Cells may require two types of signals to proliferate One signal drives cell cycle progression (mitogens) The other signal promotes cell growth Ex: pi-3 kinase pathway Learn the three main forms of cancer (carcinomas, sarcomas, leukemia) Sarcomas Tumors originating in mesenchymal tissue, such as bone, muscle or CT Rare types Carcinomas Originate in epithelial tissues, such as the cells lining the intestines, bronchi, or mammary ducts Account for 90% of all cancers Leukemia and lymphomas Lekeumia: cancer cells reside and proliferate mainly in the bloodstream rather than growing as a solid mass Often see translocation Within each of the major groups, tumors are classified by site, tissue type, histological appearance, and degree of malignancy Learn the differences between benign and malignant tumors Tumors A disease process characterized by uncontrolled cellular proliferation leading to a mass or tumor (neoplasm) Malignant tumors For a neoplasm to be cancer, it must be malignant which means its growth is no longer controlled and the tumor is capable of invading neighboring tissues or spreading (metastasizing) Cancer is a general term for malignant tumor Benign tumors Tumors that do not metastasize Instead they tend to grow in a confined local area Learn the importance of PI3-K/PTEN in growth signaling PTEN tumor suppressor important in controlling AKT growth signaling pathway The role of PTEN is to dephosphosphorylate PIP3, acting as a negative control on PKB/Akt activation Under normal growth conditions, stimulatory signals from the insulin receptor activate the enzyme phosphoinositide kinase (PI3-kinase) which phosphorylates PIP2 to generate PIP3, a lipid signaling molecule If a mutation in PTEN renders it unable to carry out its phosphatase function, PIP3 can no longer be deactivated, so continues to propagate its signal downstream Learn that cancer cells are anchorage-independent and Insensitive to population density Most normal cells need a surface in order to proliferate and survive. Tumor cells do not. Normal cells undergo apoptosis if not attached Safeguard that prevents normal cells from floating away and colonizing other tissues Cancer cells therefore exhibit anchorage-independent growth (hallmarks of cancer) Ex: E-cadherin (normally abnormally expressed in cancers) and Beta-catenin (linker protein) Cancer cells also exhibit a reduced sensitivity to density-dependent inhibition of growth Normal cells in tissue culture will usual grow until a single layer of cells is formed, then cell division stops Cancer cells grow unrestrained on top of each other Learn how cancer cells become immortalized by reactivating telomerase Normal cells divide about 50-60 times when placed in culture Stop dividing and undergo degenerative changes Cellular senescence may be due to loss of telomerase activity Cancer cells seem to proliferate indefinitely Cancer cells have telomerase activity reactivated and can maintain their telomeres. By maintaining telomere length above a critical threshold, cancer cells retain the ability to proliferate indefinitely Telomerase expression reappears in transformed cells and in many tumors An alternative mechanism for maintaining telomere length employs enzymes that exchange DNA information between chromosomes Learn about the importance of angiogenesis during oncogenesis – The roles of VEGF and matrix metalloproteinases in the process – The hazards posed by cancer cells come from uncontrolled proliferation combined with the ability to spread throughout the body – Angiogenesis (growth of blood vessels) is required for tumors to grow beyond a few millimeters in diameter – Blood vessel growth is controlled by a balance between angiogenesis activators and inhibitors Activators: VEGF, FGF These cause endothelial cells to proliferate and produce matrix metalloproteinases which breakdown extracellular matrix allowing endothelial cells to migrate MMPs are upregulated! Inhibitors: angiostatin, endostatin and thrombospondin Tumors increase production of activators and decrease inhibitors Avastin works by targeting VEGF and angiogenesis. – Tumors Increase production of activators Decrease production of inhibitors Learn about invasion and metastasis – Spreading of cancer by invasion and metastasis is a complex multistep process – Invasion Refers to the direct migration and penetration of cancer cells into neighboring tissues Take over fibroblasts and blood vessels Changes in cell adhesion, motility, and protease production allow cancer cells to invade surrounding tissues and vessels Mechanisms that allow for invasion Alteration in cell surface proteins that cause cells to adhere E-cadherin levels lower in epithelial cancers Increased motility of cancer cells Production of proteases which compromise barriers of cell movement and degrade extracellular matrix (ECM) Breaching the basal lamina in epithelial cancers Ex: plasminogen activator which converts plasminogen into plasmin Plasmin degrades ECM and basal lamina proteins Activates matrix metalloptoteinases Facilitate cell migration into blood vessel – Metastasis Able to set up colonies elsewhere in the body – very rare, which is good! Involves the ability of cancer cells to enter body fluids (blood) and travel to distant sites, where they form tumors Relatively few cancer cells survive the voyage through the bloodstream and establish metastases Either directly or indirectly through the lymph system, cancer cells arrive in the blood stream Most do not survive Those that do, usually have additional changes which make them well suited for metastasizing Blood flow patterns and organ specific factors determine where cancer cells metastasize Often metastasize the first capillary bed encountered Usually the lungs or liver Seed and soil hypothesis: some sites provide optimal growth conditions for a particular cancer cells, other sites do not Prostate cancer commonly metastasizes to bone Bone cells produce specific growth factors that stimulate prostate cells – Learn about the importance of matrix metalloproteinases in the process – Learn why metastasis occur more frequently with certain organs Learn about causes of cancer and tumor progression Cancers are caused mainly by environmental agents and lifestyle factors, most of which act by triggering DNA mutations Carcinogens Carcinogens are cancer causing agents Ex: tobacco and nasal cancer, scrotum cancer in chimney sweepers Many chemicals can cause cancer, often after metabolic activation in the liver Cytochrome p450 enzyme (Reside in SMOOTH ER) family responsible (highly polymorphic) 2 napithylamine and bladder cancer Requires conversion in active form in liver Aryl hydrocarbon hydroxylase AHH converts hydrocarbons into epoxide form that can be carcinogenic High inducibility vs low inducibility of CYPA1A1 )AHH gene) in response to smoking. Low inducibilty – less likely to develop cancer, high inducibility more likely to develop cancer The dose and the length of time exposed, increases risk Usually work by altering the DNA DNA mutations triggered by chemical carcinogens lead to cancer – how do scientists figure out if something is a carcinogen? Ames test Detect mutations that cause bacteria to regain the ability to produce histadine and grow Strain of bacteria that can’t grow in absence of histadine! Must pre-incubate the test subject with liver extract (Since it contains cytochrome p450) Cigarette smoking & lung cancer Radiation Ionization radiation increases the risk of cancer Risk is age dependent (greatest for those under 10 and elderly) Significant risk for individuals with inborn defects of DNA repair Ionizing and ultraviolet radiation also cause DNA mutations that lead to cancer UV causes thymine dimers Hepatocellular carcinoma & Aflatoxin B1 Hepatocellular carcinoma (liver cancer) The 5th most common cancer world wide In many parts of the world, hepatocellular carcinoma occurs at increased frequency because of ingestion of aflatoxin B1, a potent carcinogen produced by mold found on peanuts Aflatoxin has been shown to modify a particular base in the TP53, causing a G to T transversion in codon 249, converting an arginine codon to serine in the critically important p53 protein This mutation is found in nearly half of all hepatocellular carcinomas in patients in parts of the world in which there is a high frequency of contamination of food by aflatoxin, but it isn’t found in similar cancers in patients whose exposure to aflatoxin is low The Arg249Ser mutation in p53 enhances hepatocyte growth and interferes with the growth control and apoptosis associated with wild-type p53 LOH of TP53 in hepatocellular carcinoma is associated with a more malignant appearance of the cancer Although aflatoxin B1 alone is capable of causing hepatocellular carcinoma, it also acts synergistically with chronic hepatitis B and C infections Pathogens Viruses and other infectious agents are responsible for some cancers Oncogenic viruses 2 ways pathogens trigger cancer Virus stimulates proliferation by either introducing a viral gene or modifying host gene expression EBV, retroviruses, papillomavirus Those that cause tissue destruction and chronic inflammation. Immune response leads to the generation of free radicals and DNA damage H pylori, parasitic flatworm, hepatis B and C viruses HPV E6 & E7 Initiators vs promoters Cancer arises through a multi-step process involving initiation, promotion and tumor progression Successive rounds of random inherited change followed by natural selection During initiation, normal cells are converted to a precancerous state. Usually is a change in the DNA mutationj Promotion then stimulates the altered cells to proliferate and form tumors Then a change in behavior Chemicals that act as initiators often cause DNA damage Chemicals that act as promoters stimulate cell proliferation Natural selection will then favor cells with the fastest growth rates Tumor progression Initiation and promotion are followed by a 3rd stage, tumor progression Cancers evolve – one thing goes wrong, then another thing goes wrong, etc. Most cancers have the same origin During tumor progression, tumor cell properties gradually change as cells acquire more aberrant traits and become increasingly aggressive Any new trait which encourages growth, survival and invasiveness, usually results in those cells dominating New aberrant traits can result from epigenetic changes during tumor progression Human cancer cells are genetically unstable and frequently changing Ames test Learn that most cancers are clonal, somatic and sporadic in origin All cancers are genetic diseases The vast majority are sporadic and somatic in origin In most cancers, the mutations occur in a single somatic cell that then divides and proceeds to develop into cancer However, a single mutation is not enough to cause cancer Cancer evolves by accumulating additional genetic damage Only a small % of cancers occur as part of a hereditary cancer mutation inherited through the germ line Mutations in genes controlling proliferation and death are responsible for cancer Two distinct categories of genes involved in cancer Oncogenes (gain of function) Tumor-suppressor genes (loss of function) Learn about cancer stem cells Cancer stem cells are defined as those cancer cells that can self-renew to produce additional malignant stem cells, and at the same time generate non-tumorigenic cells such as transit amplifying cells Cancer stem cells can arise from normal stem cells that have sustained mutations to make them cancerous Cancer stem cells may arise from more differentiated cells that have undergone mutations or epigenetic changes that give them stem cell properties A small population of cancer stem cells maintain many tumors Cancer stem cells are often difficult to eradicate by conventional treatments because they replicate more slowly To cure cancer we need to find better ways to target and kill cancer stem cells Learn about the roles of oncogenes and tumor suppressors during tumorgenesis Oncogenes – are genes whose presence can trigger the development of cancer Often encode for proteins that stimulate excessive cell proliferation and or promote cell survival Nonviral oncogenesis determined by the oncogene transfection assay Oncogene transfection assay is used to analyze the ability of DNA to transform a cell Most common oncogene mutated: Ras Most oncogenes code for components of growth factor signaling pathways Tumor suppressors are genes whose loss of inactivation can lead to cancer Most common tumor suppressor mutated: p53 People who inherit one bad allele of a tumor suppressor gene are at significantly increased risk of developing cancer Require both alleles of the tumor suppressor gene to become inactivated MUST LOSE BOTH ALLELES – 2 HIT HYPOTHESIS Learn the five mechanism of converting a proto-oncogene into an oncogene Protooncogenes are normal cellular genes that can become mutated into oncogenes 5 mechanism of converting a proto-oncogene into an oncogene 1. Point mutation Ex: Ras point mutations keep it in the active form 2. Gene amplification: extra copies of genes leads to excess of protein product 3. Chromosomal translocation Ex: Burkitt’s lymphomas, CML, follicular B-cell lymphoma 4. Local DNA rearrangements Insertional mutations: retrovirus integrates in the wrong location Must know translocations cancers CML, Burkitt lymphoma & Follicular B-cell lymphoma Chronic myelogenous leukemia CML: T(9;22) (q34;q11) Philadelphia chromosome(9;22) Ph1: a translocation between chromosomes 9 and 22 Creates a fusion BCR-ABL gene at 9q and 22q Abnormal tyrosine kinase activity So treat it with a tyrosine-kinase inhibitor - Gleevac A cancer of blood cells characterized by replacement of bone marrow with malignant, leukemic cells. Many of these leukemic cells can be found circulating in the blood and can cause enlargement of the spleen, liver, and other organs. Burkitt Lymphoma T(8;14)!!!!!!!!!!!!!!!(q24;q32) Immunoglobulin put in front of the MYC gene on chromosome 8 Common tumor of children in equatorial Africa Seems to be associated with Epstein Barr Virus Translocation puts the MYC gene under the influence of an enhancer from the immunoglobulin genes Lesser likely causes: 2 with 8 lighter chains Follicular B-cell lymphoma T(14;18)(q32;q21) BCL2, an antiapoptotic gene is upregulated by a translocation which brings an immunoglobulin promoter and enhancer in proximity to the BCL2 gene Differences between gatekeeper and caretaker tumor suppressors Gatekeeper Some truly suppress tumors by regulating the cell cycle or causing growth inhibition by cell-cell contact; tumor suppressors genes of this type are gatekeepers because they regulate cell growth directly Rb and P53 (master regulators) Caretakers involved in repairing DNA damage and maintaining genomic stability Loss of both alleles of genes that are involved in repairing DNA damage or chromosome breakage leads to cancer indirectly by allowing additional secondary mutations to accumulate either in protooncogenes or in other tumor suppressors genes. Learn the two-hit origin of cancer for tumor suppressors – Both alleles of the gene need to be mutated 1st hit can be inherited 2nd hit can arise spontaneously (but once first is gone, likelihood of second being contaminated is extremely high) Epigenetic origin: gene silencing such as x inactivation and imprinting – Learn the mechanisms leading to loss of heterozygosity Individuals who have heterozygous alleles in normal tissue but have tumors that contained allels from only one of their two chromosomes LOH is the common mutational mechanism by which the remaining normal RB1 allele is lost in heterozygotes (chromosome 13 region) LOH is a feature of a number of other tumors, both sporadic and heritable, and is often considered evidence for the existence of a tumor-suppressor gene Occurs by Nondisjunction Mitotic recombination Deletion/point mutations Learn the role of epigenetics in oncogenesis Epigenetic changes that accumulate in cancer cells involve inherited chromatin structures and DNA methylation Epigenetic changes are important during the process of oncogenesis A mechanism by which developing tumor cells can inactivate tumor suppressor genes Methylation Chromatin packaging MUST know Retinoblastoma and Rb Disease caused by a mutation in a tumor suppressor gene (Rb) Diagnosis of a retinoblastoma must usually be followed by removal of the affected eye The disorder is inherited as a dominant trait, because the large number of primordial retinoblasts and their rapid rate of proliferation make it very likely that a somatic mutation will occur Penetrance is not complete. Second hit is a matter of change 60% are nonheritable (sporadic) Sporadic cases average age of onset is later than in infants with heritable form and usually develops tumor in one eye Infant with heritable retinoblastoma 400xs more likely to developing mesenchymal tumors. The risk is much higher if child received radiotherapy Get tumors in their eyeball Need to be very concerned with exposing someone to x-rays Because it causes DNA damage The sporadic version usually occurs later in life The Rb gene Involved in causing hereditary retinoblastoma Found mutated in several common adult nonhereditary cancers Lung, breast, and bladder cancer Regulates G1 to S phase transition Retinoblast protein A 110 kilodalton phosphoprotein found in many tissues The RB1 gene is a prototypical gatekeeper tumor suppressor gene Loss of Rb allows the cell to proliferate uncontrollably Rb regulates G1 to S transition It blocks cell cycle progression (G1/S phase) when it is in the hypophosphorylated state. It interacts and modifies the behavior of nuclear proteins (E2F) Dephosphorylated Rb interacts with transcription factor E2F. This prevent E2F from stimulating transcription When it is hyperphosphorylated it allows the cell to enter S phase G1 cdk-cyclin phosphorylates Rb causing it to be released from E2f and allowing e2f to stimulate transcription Must know Li Fraumeni and p53 (transcription factor) P53 is guardian of the genome A transcription factor, tumor suppressor, which influences: Cell cycle arrest – p21 Lead to apoptosis – bAX an d puma The most frequently mutated gene associated with cancer Levels usually kept really low in nucleus, with DNA damage, levels increase P21 increases A DNA-binding protein froms a tetramer Important in the cellular response to DNA damage Activates genes that stop cell division and allow DNA repair P53 also is involved with inducing apoptosis in cells that have irreparable DNA damage Loss of p53 function, allows cells with damaged DNA to survive and divide, thereby propagating potentially oncogenic mutations Li-fraumeni syndrome results from p53 mutation P53 is the most common sporadic gene associated with cancer (over half) Early onset of cancers Every generation, someone is getting affected, runs autosomal dominant in the family Inactivation of the Rb and p53 proteins is involved in the action of certain cancer viruses Human papillomavirus contains an oncogene called E7 which binds to and inactivates Rb, and another oncogene E6 which inactivates p53!!! Learn about BRCA1 & 2 in inherited breast cancer DNA repair pathway involved with: repairing double stranded DNA breaks via homologous recombination Strong genetic component Increased risk if first degree relative afflicted. More so if onset occurred prior to the age of 40 Frequent bilateral disease Less than 5% of all breast cancer BRCA1 gene: chromosome 17q21 (50% of autosomal dominant familial breast cancer) BRCA2 gene: chromosome 13q12.3 (33% of autosomal dominant familial breast cancer) Increased chance of male breast cancer (10-20% of all) Men who inherit BRCA also need to worry about breast cancer MUST KNOW Familial polyposis coli – – Colorectoal cancer: 150,000 people/year, 15% of all cancer More than one tumor – Symptoms: Little polyps on colon – Autosomal dominant – Small protion due to autosomal dominant familial polyposis coli (familial adenomatous polyposis) FAP Gardner variant is a subvariant (develop osteomas of the jaw and desmoids (tumors in the muscles of the abdominal wall) – FAP heterozygotes can have numerous benign growths by the age of 20 Usually one or more polyps become malignant Colectomy (removal of colon) prevents development of malignancies – What is wrong here? Mutation in APC – The role of APC and b-catenin Adenomatous polyposis coli = APC In wnt pathway, it’s job is to destruct b-catenin to keep levels low Linker protein with e-cadherin APC gene loci is responsible located on chromosome 5q APC encodes a cytoplasmic protein that regulates B-catenin (key target of APC) Two roles of regulates b-catenin Link the cytoplasmic portion of transmembrane cell adhesion molecules and actin cytoskeleton Activate transcription Wnt pathway Loss of APC leads to accumulation of free regulates b-catenin that is translocated to the nucleus and activates transcription Tumor suppressor APC gene mutations are common in nonhereditary forms of colon cancer Learn HNPCC and mismatch repair MICROSATELLITE INSTABILITY 2-4% of colon cancers due to HNPCC (less than 5%) An autosomal dominant disease onset during early adulthood without the adenomatous polyps of FAP Heterozygous males have 90% lifetime risk of developing cancer Female heterozygotes have 70% chance, 40% risk of endometrial cancer and 1020% risk for cancer of bilary or urinary tract, and the ovary DNA isolated from tumors display microsatellite instability Symptoms Abdominal pain Anemic Blood in stool Diarrhea, trouble going to the bathroom HNPCC is caused by mutations in DNA repair genes HNPCC is a group of 5 similar syndromes caused by mutations in 1 of 5 distinct DNA repair genes Inherited defects in genes required for mismatch repair are responsible for HNPCC The genes: MLH1, MSH2, PMSL1, PMSL2, and MSH6 MLH1 and MSH2 together account for 60-70% of HNPCC The HNPCC genes are prototypical caretaker tumor-suppressor genes Multi hit model of colon cancer induction Usually more than one pathway is disrupted Colon cancer evolves through distinct, well-characterized morphological stages Intermediate stages: polyps, benign adenomas, and carcinomas can be isolated Mutations in each of the morphological stages can be identified Resulting in a series of mutations that commonly arise in a well-defined order, hence multi-hit model Learn about microRNAs (miRNAs) and cancer The catalogue of genes involved in cancer also includes genes that are transcribed into noncoding RNAs from which regulatory microRNAs (miRNAs) are generated There are at least 250 miRNAs in the human genome that carry out RNAmediated inhibition of the expression of their target protein-coding genes, either by inducing the degradation of their targets’ mRNA or by blocking their translation Approx. 10% of mRNAs have been found to be either greatly overexpressed of down-regulated in various tumors, sometimes strikingly so, and are referred to as ONCOMIRS One example is the 100-fold overexpression of the miRNA miR-21 in glioblastoma multiforme, a highly malignant form of brain cancer Oncomir = oncogenic micro RNA Overexpression of some miRNAs can suppress the expression of tumorsuppressor gene targets Whereas loss of function of other miRNAs may allow overexpression of the oncogenes they regulate Since each miRNA may regulate as many as 200 different gene targets, overexpression or loss of function of miRNAs may have widespread ocogenic effects because many genes will be dysregulated RB and Oncomir miR-106a The retinoblastoma gene RB1 is frequently mutated in many cancers, including breast cancer Ex. 13q14 LOH observed in human breast cancers is associated with loss of RB1 mRNA in the tumor tissues In still other cancers, the RB1gene is intact and its mRNA appears to be at or near normal levels, and yet the p110 Rb1 protein is deficient This anomaly has now been explained by the recognition that RB1 can be down-regulated in association with overexpression of the oncomir mi-RNA 106a, which targets RB1 mRNA and blocks its translation Thus, miR-106a might be considered an oncogene that exerts its effect by reducing the expression of the TSG encoding the p11- Rb1 protein One group blocks tumor suppressors Too much of them Other group target protooncogenes Learn about genetic instability and gene amplification Genetic instability Trait of cancer cells in which abnormally high mutation rates are caused by defects in DNA repair and/or chromosomes sorting mechanisms Inherited defects in genes required for mismatch repair are responsible for HNPCC Xeroderma pigmentosum caused by inherited defects in UV damage excision repair pathway Hereditary breast cancers involves mutated forms of BRCA1 or BRCA2, which are involved in repairing double-strand breaks within DNA Genetic instability can cause defects in mitosis and aneuploidy Some cancer cells have an extra chromosome Tumors generally have chromosome and genome mutations that reflect defects either in double-stranded break repair (generate chromosomal translocations) or maintenance of the fidelity of how chromosomes align on the mitotic spindle during mitosis (lead to nondisjunction and aneuploidy) Gene amplification What are usually amplified? Protooncogenes and tumor suppressors Many additional copies of a segment of genome present in cell Common in many cancers – more amplification that you see, the worse the prognosis Amplified segments detect by CGH typically as 2 types of cytogenetic changes Double minutes; very small accessory chromosomes Homologous staining regions (HSR) that do not band normally and contain multiple, amplified copies of a particular DNA segment Amplified regions are known to contain extra copies of proto-oncogenes (myc, Ras, EGFR) Ex. Amplification of mycn proto-oncogene for n-myc is a clinical indicator for prognosis in the childhood cancer, neuroblatoma Myc-n is amplified more than 200xs in 40% of advanced stages of neuroblastoma Learn about the hallmarks of cancer 1. Self sufficiency in growth signals Do not need to be stimulated to grow Ras mutation 2. Insensitivity to antigrowth signals Mutations involving RB or TGFbeta/smad pathway 3. Evasion of apoptosis Mutations in p53 or bcl2 which promote cell survival 4. Limitless replicative potential Mutation allows cell to maintain telomere length 5. Sustained angiogenesis Blood supply and nutrients re vital for tumor growth 6. Tissue invasion and metastasis Decrease in cell adhesion (E-cadherin mutation) and/or increase in protease production 7. Cancers induce help from normal stromal cells in their local environment 8. They are defective in the intercellular control mechanisms that normally stop cell divisions permanently in response to stress (such as hypoxia) or DNA damage An enabling trait: genetic instability Allows for generation of genetic mutations to occur Introduction about diagnosis, screening and treatment of cancer – Know Herceptin & breast/ovarian cancer, Gleevec & CML, and Avastin – Diagnosis Cancer is diagnosed by microscopic examination of tissue specimens Biopsy of tumor for microscopic analysis Is a sufficient numver of these traits are observed, it can be concluded that cancer is present, even if invasion and metastasis have not yet occurred Tumor grading: numerical grade based on microscopic appearance Lower numerical grades are assigned to tumors who cells exhibit normal differentiated features and display modest abnormalities The highest grade are anaplastic: poorly differentiated, abnormal in appearance. They bear no resemblance to the cells of the tissue they arose from – Screening techniques for cancer Higher cure rates when cancer is treated before it spreads Important to screen and detect cancers as early as possible Pap smear: technique for early detection of cervical cancer. Analyze sample of vaginal secretions for abnormal cells Large irregular nuclei or prominent variations in cell size or shape Can detect before metastasis Prevent many cancer deaths Mammography for detecting breast cancer Colonoscopy for detecting colon cancer PSA test: measure prostate-specific antigen in the blood High PSA indicates prostate problem Proteomic analysis of protein in the blood Utilize mass spectrometry – Used for detecting ovarian cancer Screens for 5 protein markers of ovarian cancer Treatments for cancer Surgery radiation, and chemotherapy Radiation therapy employs x-rays or other types of radiation to kill cancer cells by 2 mechanisms Radiation activates p53 pathway leading to apoptosis Severly destroys DNA and prevent progression through mitosis Chemotherapy to kill dividing cells, four categories Antimetabolites Inhibit DNA synthesis Methotrexate, fluorouracil, and mercaptourine Alkylating agents Inhibit DNA function by crosslink DNA double helix Cyclophosphamide, cholrambucil, and cysplatin Antibiotics Inhibit DNA function by binding DNA or topoisomerases Bleomycin, docorubicin Plant-derived drugs Either inhibit topoisomerase or disrupt microtubules Ectoposide and taxol-binds and stabilizes (vinlblastin, vinchristin) Block action of hormone require growth: tamoxifen for breast cancer Magic bullet strategy: selectively destroy the cancer cells and not the normal cells Immunotherapies explot the ability of the immune system to recognize cancer cells Tumors occasionally regress in people who develop bacterial infections because infections trigger immune response Bacilis calmette-guerin bacteria is inserted into the bladdter after primary tumor is removed Drugs: interferon alpha and interleukin 2 Herceptin and gleevec are cancer drugs that act through molecular targeting Gleevec (imatinib) is a small molecule that binds to and inhibits the tyrosine kinase activity or BCR-ABL in chronic myelogenous leukemia CML (Philadelphia chromosome) How gleevac blocks the activity of bcr-abl protein and halts CML -Sits in the ATP-binding pocket of the tyrosine residue in a substrate protein This blocks onward transmission of a signal for cell proliferation and survival Herceptin is an antibody which recognizes the ERBB2 growth factor receptor Breast and ovarian cancers – Anti angiogenic therapies act by attacking a tumor’s blood supply. Block blood vessel formation in tumors. Avastin – is a monoclonal antibody that binds to and inactivates VEGF Best way to treat cancer and aids Multiprong attack Personalized medicine