Overview:
advertisement

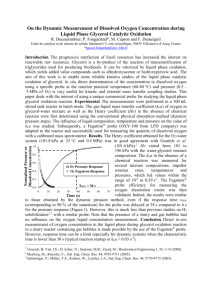
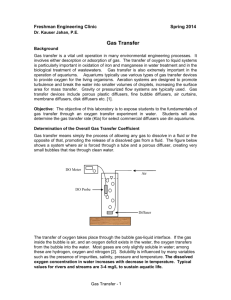
Measurement of KLa in a Sparged Bioreactor Overview: In these experiments we will: 1) Model the process for raising the level of dissolved oxygen in a tank fermenter type bioreactor. This process involves bubbling air or oxygen into a fermenter with stirring. Oxygen is transferred from the gas phase (bubbles) to the liquid phase (in our case waterin an actual process, fermentation broth). The two parameters of interest will be the probe dynamics (1st order time constant, τ) and the KLA* (volume averaged mass transfer coefficient lumped with total interfacial area for mass transfer). 2) We will use a dissolved oxygen (D.O.) probe to measure the concentration of oxygen in the liquid phase. The probe does not respond immediately to changes in liquid concentration. Therefore we first need to determine the probe dynamics for a dissolved oxygen (D.O.) probe. We will do this by first performing a “step change” experiment to determine τ (we actually also observe what appears to be a delay time in the probe response which we include in the model also). 3) In the second series of experiments we will measure the apparent rate of oxygen mass transfer from gas to liquid phase in a bioreactor using the DO probe. We will then use #1 and #2 to determine KLA as a function of both the rate of power input from stirring and the rate of “sparging” gas into the bioreactor. Procedure 1) Familiarize yourself with the bioreactor, D.O. measurement system and spreadsheet/ VBA code for data acquisition and curve fitting to the process model. The manual for the bioreactor, as well as dissolved oxygen tables are in the drawers on top of the unit. The data acquisition system uses VBA code to import data directly into Excel. The D.O. meter puts out a current of 0 to 20ma. We have a resister wired across the outputs so that this results in a voltage of approximately 0 volts at zero dissolved oxygen content, and 5 volts at 100% saturation. The dissolved oxygen tables will tell you how many mg/ml (ppm) to expect at 100% saturation depending on temperature. Look at the “model” cells. Convince yourself that this model is the same one developed in class and the Brown article. The only difference is that we are describing the probe * where kL is the oxygen transport coefficient (cm3/h), a is the gas-liquid interfacial area (m2/m3), kLa the average volumetric oxygen transfer coefficient (h-1), C* is saturated DO concentration (mg/l) (approx. 8 mg/l at 25 deg. C and 1 atm.), CL is the actual DO concentration in the liquid (mg/l), and N O2 is the rate of oxygen transfer (mg O2/ l h) (Shuler and Kargi, 1992) response as “first order with time delay” which fits our D.O. probe very well and does not significantly change the model response- only the starting point. 2) Perform a “step change” experiment: The fermenter should be sparged with oxygen and about 5 L/min and a stirring rate of 500 RPM until the concentration reaches a maximum at the saturation value. Very carefully place the probe in a stirred beaker containing sodium sulfite. The sodium sulfite reacts quickly with any oxygen entering the liquid phase and keeps the concentration at essentially zero. Note that bumping the tip of the probe on the beaker or fermenter may damage the probe membrane and/or the probe tip! Observe the oxygen concentration drop to zero. When the concentration reading is stable you are ready to do the step change. Set the value of KLa in the spreadsheet equal to 100000 (this is the same saying that the rate of mass transfer is on the order of 0.00001 seconds- essentially infinitely fast compared to the probe constant we will calculate from our data. Convince yourself that by setting the KLa so high, one of the model terms drops out so that we can determine directly. An ideal step change is the same as infinitely fast mass transfer: we are going from zero to 100% concentration “instantaneously”. Again very carefully but quickly, move the probe from the beaker to the fermenter. You should place the probe straight down into the solution to avoid hitting the impellor blades and tighten the collar on the probe mount. At the same time the probe enters the solution, start the data acquisition by pressing “start” on the spreadsheet interface. You should observe a delay followed by a probe response that starts quickly and then asymptotically approaches the saturated solution concentration in the fermenter. There are several ways to determine the probe response parameters. The easiest is to observe where the increase in oxygen starts and to enter this value as the “time delay”. The Solver function in Excel may then be used to change the value of until the sum square of the errors between the model equation and the data is minimized. Alternatively you could observe the initial slope of the response and calculate the value of 0.632 of the full response (C/C* = 0.632). If you use Solver, check to see that the value of is very close to 0.632 of the response. 3) Use “Gassing in” Experiments to determine: a) KLa (e, vs) using the form: K L a ae b vsc (1) Where e is the power input per unit volume (you will need to refer to the on-line and suggested references to find this value as a function of stir rate and gas holdup) and vs is the superficial velocity of sparging gas through the fermenter. The use of these variables creates a somewhat general expression, although the equation still depends on the specific size and configuration of the fermenter, especially the leading parameter “a”. The parameters “a”, “b” and “c” may be determined by a non-linear curve fitting regression) program such as that found in Polymath. And: b) Determine KLa (N,Q) using a factorial method to determine the dependence of KLa for the specific conditions found in our fermenter, as well as any interaction between Q (volumetric flow rate) and N (RPM). (To use this method we will need to reverse engineer the factorial design by going to the “historical data” section of the factorial design in the Design Expert software. Since we have already done the experiment as a full factorial design) For the experiment you will do 16 runs in random order for the gassing in method. Choose 4 stir rates between 300 and 900 RPM and 4 volumetric gas flow rates from 2 to 12 liters per minute oxygen†. Doing all possible combinations at 4 levels is a 42 full factorial method: 2 factors (variables) at 4 levels each. Gassing in method: Once you have entered values for the time delay and time constant, strip any dissolved oxygen from the fermenter by sparging with nitrogen. To speed this process up, use a high stir speed of abut 900 and a sparged rate of about 15 L/min. Once the D.O. concentration stabilizes zero, set the conditions (N,Q) to those listed in the randomized design for run 1. Second stage pressure readings on both the oxygen and nitrogen tanks † Also do one run for intermediate conditions using house air, to see if this significantly affects the value of KLa. should be 20 psi so that when you switch to oxygen the flow rate will stay at these set values. Switch the inlet valve from nitrogen to oxygen and simultaneously start the data acquisition spreadsheet by pressing “start” on the screen. You should again observe a delay of a few seconds, followed by what appears to be a second order response (sigmoidal shape) that asymptotically approaches the saturation concentration. The data will be plotted in blue, and the model line in pink. After the measured concentration stabilizes at the maximum value (get close, but you do not need to run for a prolonged period of time where the time of the run is more of the stable period than the dynamic one. . . .we want to determine the dynamics, not any small offset from our predicted C*) End the run by repeatedly hitting “escape” on the keyboard, or the mouse button. This will allow you to stop the run-time program. You may determine KLa for this run while setting up for the next- this should be very time-efficient for you. To find KLa, again go to Solver. Now, the values for the probe response are fixed and the only variable in our model is the KLa value. Fit the model to the data by having Solver to change the value of KLa, optimizing so that the SSE (sum of squares of the difference between each data point and model point) is minimized. The model should fit the data closely, and the value of KLa may be recorded as the result for those specific conditions. Save your time vs. concentration data in a different spreadsheet for future reference. Repeat for all 16 runs. Starting References‡: 1. Handouts in class, especially the article by Brown 2. Other materials on the Lab Manuals Web Page: http://www.eng.auburn.edu/~drmills/mans486/manindex4860new_page2.html 3. Linek, V, Sinkule, J, and Benes, P, 1991, Critical Assessment of Gassing-In Methods for measuring kLa in Fermentors, Biotechnology and Bioenginering 38, 323-330 4. Perry’s Handbook, 50th ed., section 28 5. Shuler, ML and Kargi, F., 1987, Bioprocess Engineering: Basic Concepts, Prentice Hall, NJ ‡ Also any other good biochemical engineering text (i.e. Bailey and Ollis) , Perry’s Handbook (Liquid-Gas Systems, Biochemical Engineering).