28-Fura Suspension Protocol
advertisement

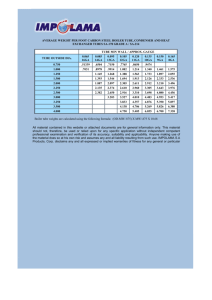
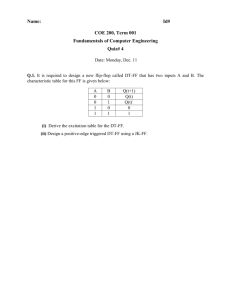
Fura-2-AM: Ca2+Measurement for Cells in Suspension - Ignite fluorimeter lamp. Wait 5 minutes. - Switch-on PTI water-bath so it can warm to 370C. Pre-warm all solutions/buffers to 370C before using. 1. Grow 10 cm plate of cells until they are confluent (1 to 2 x 107 cells). 2. Wash 2 times with PBS. 3. Add 1.5 mL of trypsin to each plate and incubate at 370C for ~ eight minutes. 4. Add 8.5 mL DMEM (no FBS) to neutralize trypsin. 5. Transfer contents of each plate to an individual 15 mL falcon tube. 6. Spin tube at 800 rpm (S4180 rotor) for 5 minutes at room temperature. 7. Re-suspend cells by adding 10 mL Ca2+-free buffer to each tube. 8. Spin tube at 800 rpm for 5 minutes at RT. 9. Re-suspend pellet in 5 mL Ca2+-free buffer. 10. Aliquot 10 L and count cells with hemacytometer. 11. Count 4 different regions of 16 squares and determine average number of cells. (Total # Cells in 1 mL = Average x 104 x 2mL) * You need to fura load 1 x 107 cells. (LOADING VARIES WITH CELL TYPE) 12. Spin tube at 800 rpm for 5 minutes at RT. 13. Re-suspend pellet in 1 mL Ca2+-free buffer. 14. In an eppendorf tube put: 10 L of 10% BSA (final conc. 0.1%), 2 L of 10% Pluronic F-127 (final conc. 0.02%), and 3 L of 1 mM Fura-2-AM (final conc. 3 M). 15. Add the 1 mL of cells to the eppendorf tube with the Fura mixture. 16. Cover eppendorf tube with foil and incubate in 370C H2O bath for 30 minutes. 17. Invert tube a few times every ~ 8 minutes during the incubation period. - Switch-on all components of PTI fluorimeter. - Adjust all six fluorimeter slits to 2 nm. 18. After incubation period, spin tube at 900 rpm (switch to smaller F2402H rotor) for 5 minutes at RT. 19. Discard supernant. 20. Re-suspend precipitate in 1 mL Ca2+-free buffer. Foil wrap tube and use cells immediately. 21. Aliquot 140 L of Fura-loaded cells (i.e. 1.4 x 106 cells) into “small” quartz cuvette with 1260 L Ca2+-free buffer. 22. The stock Fura-loaded cells can be placed in 370C H2O bath. Acquire your data. Excitation Scan Measurement for Fura-2-AM 2 excitation wavelengths (i.e. range) with emission wavelength fixed. 1. Turn-off ALL equipment around PTI instrument. 2. Follow the “GETTING STARTED” steps. 3. Open the FeliX software. 4. In FeliX, select “Acquire” and then “Excitation Scan.” 5. The software will automatically adjust the instrument. 6. Confirm that the excitation and emission nm settings reported by the software agree with that of the instrument + 1 nm (i.e. check the “counters”). 7. Enter parameters: Excitation Emission: Step Size: Integration Time: Averages: Start: 300 nm 510 nm 0.5 nm 0.25 sec. 1 Stop: 450 nm 8. Set all slits to 2 nm bandpass (i.e. 1.0 turns). --- There are six slits to adjust. 9. Put cuvette + sample into instrument. 10. Now select “ACQUIRE (PREP)”. 11. Select “Start” to acquire data. 12. Add drugs/solutions at the appropriate time (use clock in window at top left of FeliX) by selecting “PAUSE”, opening the fluorimeter lid, and injecting solution. Click on “CONTINUE” to resume data acquisition. Press “space-bar” to insert event marker. Excitation Ratio Measurement for Fura-2-AM 2 excitation wavelengths are measured with emission wavelength fixed. 1. Turn-off ALL equipment around PTI instrument. 2. Follow the “GETTING STARTED” steps. 3. Open the FeliX software. 4. In FeliX, select “Acquire” and then “Excitation Ratio.” 5. The software will automatically adjust the instrument. 6. Confirm that the excitation and emission nm settings reported by the software agree with that of the instrument + 1 nm (i.e. check the “counters”). Enter parameters: Emission: Integration: Duration: Repeats: Pause: View Window: Excitation 1: 340 nm 510 nm 0.1 sec 6000 sec 0 Excitation 2: 380 nm 6000 sec 7. Select “Display”. 8. Highlight Emitter A curve (numbered 1 on left side); now, in the right corner click of this window click on “Derived Data” and select “:1 new”. Do the same for the other Emitter A curve (number 3 on the left side). 9. Highlight Emitter B curve (numbered 2 on left side); now, in the right corner click of this window click on “Derived Data” and select “:2 new”. Do the same for the other Emitter B curve (number 4 on the left side). 10. In the center of this window you see 4 bullet holes. Select the hole beside number 1. 11. In the lower left, you will see Source boxes. For Source 1 select “1”. For Source 2 select “3”. 12. In the lower, center portion of this window, click on “Derived Data” and select “Source1/Source2”. 13. Now in the lower right corner, under “Derived Data” select “:3 new”. 14. In the center of the window are the 4 bullet holes; Select the hole beside number 2. 15. In the lower left, you will see Source boxes. For Source 1 select “2”. For Source 2 select “4”. 16. In the lower, center portion of this window, click on “Derived Data” and select “Source1/Source2”. 17. Now in the lower right corner, under “Derived Data” select “:4 new”. 18. Be certain that all slits have been set to 2 nm bandpass (i.e. 1.0 turns). --- There are six slits to adjust. 19. Put cuvette + sample into instrument. 20. Now select “ACQUIRE (PREP)”. 21. Select “Start” to acquire data. 22. Wait for 340 nm and 380 nm baselines to stabilize (~ 8 minutes). 23. Add drugs/solutions at the appropriate time (use clock in window at top left of FeliX) by selecting “PAUSE”, opening the fluorimeter lid, and injecting solution. Click on “CONTINUE” to resume data acquisition. Press the spacebar to deposit an event marker. Time 2 minutes 3 minutes 5 minutes 10 minutes 15 minutes (Add as mixture) Time 2 minutes 3 minutes 5 minutes 10 minutes 15 minutes (Add as mixture) Drug Volume 7.5 L 7.5 L 7.5 L 7.5 L 3 L 3 L 4.5 L 3 L Drug Volume 7.5 L 7.5 L 7.5 L 7.5 L 3 L 3 L 4.5 L 3 L Bradykinin Bradykinin Bradykinin Thapsigargin A23187 Ionomycin 1M CaCl2 50 mM ATP Bradykinin Bradykinin Bradykinin Thapsigargin A23187 Ionomycin 1M CaCl2 50 mM ATP * press the spacebar to deposit an event marker.