(2) Book Chapter Publications as an Assistant Professor and an
advertisement

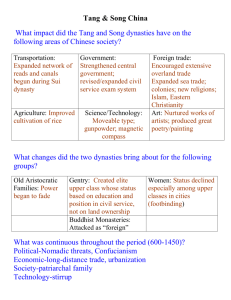
Tang Updated CV CURRICULUM VITAE Guiliang Tang, Associate Professor (Nov 2011-present), Department of Biological Sciences, Michigan Technological University, Houghton, MI 49931 EDUCATION Ph.D., The Weizmann Institute of Sciences, Rehovot, Israel, 2001 M.Sc., Anhui Agricultural University, Hefei, China, 1991 B.Sc., Anhui Agricultural University, Hefei, China, 1983 RESEARCH INTERESTS My research interests focus on: 1) Biotechnology and Natural Products, 2) Gene Silencing, and 3) RNA interference (RNAi) and microRNA (miRNA). PROFESSIONAL EXPERIENCE 2011-present Associate Professor, Department of Biological Sciences, Michigan Technological University, Houghton, MI 2011-2011 Associate Professor (Tenured), University of Kentucky, Lexington, KY 2005-2011 Assistant Professor (Tenure-track), University of Kentucky, Lexington, KY 2001-2005 Postdoctoral Research Fellow, Department of Biochemistry and Molecular Biology at the University of Massachusetts Medical School, Worcester, MA 1995-2001 Graduate research assistant, The Weizmann Institute of Science, Israel. 1991-1995 Assistant Professor, College of Life Science, Anhui Agricultural University, Hefei, China 1988-1991 Graduate research assistant, College of Life Science, Anhui Agricultural University, Hefei, China 1983-1988 Research Assistant, Guichi Agricutural Research Center, Anhui, China 1 Tang Updated CV EXTRAMURAL FUNDING Summary Funds allocated to my program since July 2005: $3,584,854 National funding (NSF & USDA-NRI): $3,129,854 Kentucky funding (KSEF): $250,000 University funding (KTRDC): $175,000 Industry funding (P&G): $30,000 National-Competitive Grants ($3,129,854) Guiliang Tang (PI), Hairong Wei (co-PI), Wenbo Ma (co-PI), Xuemei Chen (coPI), and Harold Trick (co-PI) (2014-2017) NSF-PGRP: Targeting microRNAs for Destruction in Crops by Short Tandem Target Mimic (STTM). NSF, $2,499,979.00. Guiliang Tang (PI) (2010-2014) NSF-EAGER: Random RNAi to Generate Dominant Mutant Pools for Gene Discovery. NSF, $ $295,576. Guiliang Tang (PI), 2006-2009, Developing microRNA vectors for gene suppression, United States Department of Agriculture-National Research Initiative (USDA-NRI) Competitive Grants Program, $167,278 Guiliang Tang (PI), 2006-2009, Random RNAi screening for negative regulators of drought resistance in Arabidopsis, USDA-NRI Competitive Grants Program, $100,000 Xuemei Chen (PI) and Guiliang Tang (Co-PI), 2007-2010, Role of methylation in microRNA metabolism in Arabidopsis. National Science Foundation (NSF), $500,000 ($67,000 was allocated to my research program) Kentucky Competitive Grants ($250,000) Guiliang Tang (PI), 2008-2011, MicroRNA array technology and microRNA biomarkers for various human diseases. Kentucky Science and Engineering Foundation (KSEF), $150,000. Guiliang Tang (PI), 2008-2011, Developing a novel poly-cistronic (poly-cis) microRNA vector to silence multi-genes in plants and the application in lignin biosynthesis. Kentucky Tobacco Research and Development Center (KTRDC), $100,000 University Competitive Grants ($175,000) Guiliang Tang (PI), 2005-2006, Developing microRNA vectors for gene suppression in plants (KTRDC), $50,000 Ling Yuan (co-PI), Jan Smalle (co-PI), Guiliang Tang (co-PI), Hongyan Zhu (co-PI). (2009-2011) Joint Initiative on Advanced Plant Bioengineering, Kentucky Tobacco Research and Development Center, 2008-2010; $500,000. ($125,000 was allotted to my research program) 2 Tang Updated CV Other Grants ($30,000) George J. Wagner (PI), Joseph Chappell (PI), Guiliang Tang (co-PI) and Ling Yuan (co-PI), 2008-2011, Production of abietic acid and other potentially-useful, related diterpenoids in tobacco trichomes, Procter and Gamble (P&G), $532,321. ($30,000 was allotted to my research program) PUBLICATIONS (1) Journal Research Publications as an Assistant Professor and an Associate Professor at the University of Kentucky and Michigan Technological University (* corresponding author; 1 postdoc and 2 student under my supervision) 1. Sachin Teotia and Guiliang Tang (2015) To bloom or not to bloom: Role of microRNAs in plant flowering. Molecular Plants. (In press) 2. Xiaoyun Jia, Na Ding, Weixin Fan, Jun Yan, Yiyou Gu, Xiaoqing Tang, Runzhi Li, and Guiliang Tang (2015) Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Science. (In press) 3. Peng T, Sun H, Qiao M, Zhao Y, Du Y, Zhang J, Li J, Tang G, Zhao Q. (2014) Differentially expressed microRNA cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice. BMC Plant Biol. 2014 Jul 23;14(1):196. 4. Zhenying Cai, Jingjing Liu, Haijiao Wang, Cangjing Yang, Yuxiao Chen, Yongchi Li, Shangjin Pan, Rui Dong, Guiliang Tang, Juan de Dios Barajas-Lopez, Hiroaki Fujii, and Xuelu Wang (2014) GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci U S A. 2014 Jun 13. pii: 201316717. 5. 2.James Wong, Lei Gao, Yang Yang, Jixian Zhai, Siwaret Arikit, Yu Yu, Shuyi Duan, Vicky Chan, Qin Xiong, Jun Yan, Shengben Li, Renyi Liu, Yuanchao Wang, Guiliang Tang, Blake C. Meyers, Xuemei Chen and Wenbo Ma (2014) Roles of Small RNAs in Soybean Defense against Phytophthora sojae Infection. Plant J. 2014 Jun 18. doi: 10.1111/tpj.12590. 6. Ming Chi, Basdeo Bhagwat, W David Lane, Guiliang Tang, Yinquan Su, Runcang Sun, B Dave Oomah, Paul A Wiersma, Yu Xiang (2014) Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biology 14: 62 7. Bhagwata B, Chia M, Su L, Tang H, Tang G.*, Xiang Y. (2013) An in vivo Transient Expression System Can Be Applied for Rapid and Effective Selection of Artificial microRNA Constructs for Plant Stable Genetic Transformation. Journal of Genetics and Genomics 40(5): 261-70. 8. Dianwei Han, Guiliang Tang*, and Jun Zhang (2013) A Parallel Strategy for Predicting the Secondary Structure of Polycistronic MicroRNAs. Int. J. Bioinformatics Research and Applications. 9(2): 134-55. 9. Jun Yan1, Yiyou Gu2, Xiaoyun Jia2, Wenjun Kang2, Shangjin Pan2, Xiaoqing Tang, Xuemei Chen and Guiliang Tang* (2012) Effective Small RNA Destruction by the Expression of a Short Tandem Target Mimic in Arabidopsis thaliana. The Plant Cell 24(2): 415-27. 3 Tang Updated CV 10. Guiliang Tang*; Jun Yan1, Yiyou Gu2, Mengmeng Qiao2, Ruiwen Fan, Yiping Mao, Xiaoqing Tang* (2012) Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58(2):118-25. 11. Ren L.2 and Tang G.* (2012) Identification of sucrose-responsive microRNAs reveals sucrose-regulated copper accumulations in an SPL7-dependent and independent manner in Arabidopsis thaliana. Plant Science, 187: 59-68 12. Dianwei Han, Jun Zhang*, and Guiliang Tang* (2012) MicroRNAfold: premicroRNA secondary structure prediction based on Modified NCM model with thermodynamics-based scoring strategy. International Journal of Data Mining and Bioinformatics 6(3): 272-91. 13. Iyer NJ, Jia X, Sunkar R, Tang G, Mahalingam R*. (2012) microRNAs responsive to ozone-induced oxidative stress in Arabidopsis thaliana. Plant Signal Behav 7(4): 484 - 491. 14. Lijuan Ji, Xigang Liu, Jun Yan2, Wenming Wang, Rae Eden Yumul1, Yu Ju Kim, Thanh Theresa Dinh, Jun Liu, Xia Cui, Binglian Zheng, Manu Agarwal, Chunyan Liu, Xiaofeng Cao, Guiliang Tang, and Xuemei Chen* (2011) ARGONAUTE10 and ARGONAUTE1 Regulate the Termination of Floral Stem Cells through Two microRNAs in Arabidopsis. PLoS Genet. 7(3):e1001358. Epub 2011 Mar 31. 15. Wang-Xia Wang, Bernard R. Wilfred, Sindhu K. Madathil, Guiliang Tang, Yanling Hu, James Dimayuga, Arnold J. Stromberg, Qingwei Huang, Kathryn E. Saatman, and Peter T. Nelson* (2010) MiR-107 regulates Granulin/Progranulin with implications for traumatic brain injury and neurodegenerative disease. American Journal of Pathology 177(1): 334-45 16. Mian Gu, Ke Xu, Aiqun Chen, Yiyong Zhu, Guiliang Tang, and Guohua Xu* (2010) Expression Analysis Suggests Potential Roles of microRNAs for Phosphate and Arbuscular Mycorrhizal Signaling in Solanum lycopersicum. Physiol Plant. 138(2): 226-37 17. Xiaoyun Jia2, Wang-Xia Wang1, Ligang Ren2, Qi-Jun Chen1, Venugopal Mendu2, Benjamin Willcut2, Randy Dinkins, Xiaoqing Tang1, and Guiliang Tang* (2009) Differential and dynamic regulation of miR398 and its targets in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Molecular Biology. 71: 51-59. 18. Xiaoyun Jia2, Ligang Ren2, Qi-Jun Chen1, Runzhi Li and Guiliang Tang* (2009) UV-B responsive microRNAs in Populus tremula. J. of Plant Physiology 166: 20462057. 19. Xiaoqing Tang1, Latha Muniappan, Guiliang Tang*, and Sabire Ozcan* (2009) Identification of glucose-regulated miRNAs from pancreatic beta cells reveals a role for miR-30d in insulin transcription. RNA.15: 287-293. 20. Wang-Xia Wang, Bernard W. Rajeev, Arnold Stromberg, Na Ren, Guiliang Tang, Qingwei Huang, Isidore Rigoutsos, and Peter T. Nelson* (2008) The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of BACE1. Journal of Neuroscience 28(5): 1213-1223 21. Xiaoqing Tang1, Jozsef Gal, Xun Zhuang2, Wangxia Wang1, Haining Zhu, and Guiliang Tang* (2007) A simple array platform for microRNA analysis and its application in mouse tissues. RNA 13: 1803-1822 4 Tang Updated CV (2) Journal Review Publications as an Assistant Professor and an Associate Professor at the University of Kentucky and Michigan Technological University (* corresponding author; 1 postdoc and 2 student under my supervision) 22. Tang G.* and Tang X. * (2013) Short Tandem Target Mimic: A Long Journey to the Engineered Molecular Landmine for Selective Destruction/Blockage of microRNAs in Plants and Animals. Journal of Genetics and Genomics. 40 (6): 291–296. 23. Xiaoyun Jia2 and Guiliang Tang* (2011) MicroRNA-mediated DNA methylation in plants. Frontiers in Biology 6(2): 133–139 24. Guiliang Tang* (2010) Plant microRNA: An insight into the gene structure and evolution. Seminars in Cell and Developmental Biology 21: 782-789 25. Xiaoqing Tang1*, Guiliang Tang, and Sabire Ozcan (2008) Role of MicroRNAs in Diabetes. BBA-Gene Regulatory Mechanisms 1779: 697-701 26. Guiliang Tang*, Xiaoqing Tang1, Venugopal Mendu2, Xiaohu Tang1, Xiaoyun Jia2, Qi-Jun Chen1, and Lihen He2 (2008) The art of microRNA: various strategies leading to gene silencing via an ancient pathway. BBA-Gene Regulatory Mechanisms 1779: 655-662 27. Peter T. Nelson*, Wang-Xia Wang, Bernard R. Wilfred, Guiliang Tang (2008) Technical variables in high-throughput miRNA expression profiling: Much work remains to be done. BBA-Gene Regulatory Mechanisms 1779: 758-765 28. Guiliang Tang*, Gad Galili and Xun Zhuang2 (2007) RNAi and microRNA: Breakthrough technologies for the improvement of plant nutritional value and metabolic engineering. Metabolomics.3: 357-369 (2) Book Chapter Publications as an Assistant Professor and an Associate Professor at the University of Kentucky and Michigan Technological University (* corresponding author; 1 postdoc and 2 student under my supervision) 29. Xiaoqing Tang1, Xiaohu Tang1, Jozsef Gal, Natasha Kyprianou, Haining Zhu, and Guiliang Tang* (2010) Detection of microRNAs in prostate cancer cells by microRNA array, Methods Mol Biol. 2011;732: 69-88. 30. Xiaoyun Jian2, Venugopal Mendu2, and Guiliang Tang* (2010) An array platform for identification of stress-responsive miRNAs in plants, "Methods in Molecular Biology; Plant Stress Tolerance- Methods and Protocols" Methods Mol Biol. 639: 253-69 31. Ricky Lewis, Venugopal Mendu2, David McNear, and Guiliang Tang* (2009) Roles of microRNAs in Plant Abiotic Stress, Molecular Techniques in Crop Improvement. 2nd Edition, edited by S. Mohan Jain and D.S. Brar. Springer Netherlands, pp 357372. 32. Wang-Xia Wang1, Peter Nelson, and Guiliang Tang* (2008) RNA Interference, mechanisms and proteins involved in. Wiley Encyclopedia of Chemical Biology. Wiley Encyclopedia of Chemical Biology. 33. Guiliang Tang*, Yu Xiang, Zhensheng Kang, Venugopal Mendu2, Xiaohu Tang1, Xiaoyun Jia2, Qi-Jun Chen1, and Xiaoqing Tang1 (2008) Small RNA technologies: 5 Tang Updated CV siRNA, miRNA, antagomiR, target mimicry, miRNA sponge and miRNA profiling. Current Research and Perspectives in MicroRNAs. Springer Netherlands, pp 17-33 34. Wang-Xia Wang1, Bobby Gaffney, Arthur G. Hunt and Guiliang Tang* (2007) microRNAs and Plant Development. Encyclopedia of Life Sciences. 35. Yu Xiang and Guiliang Tang*. (2006) RISC Biology, “microRNA: Biology, Function and Expression” edited by Clarke, N.J and Sanseau, P, DNA Press. pp 2969. (3) Proceeding Publications as an Assistant Professor and Associate Professor at the University of Kentucky and Michigan Technological University 36. Dianwei Han, Guiliang Tang*, and Jun Zhang* (2009) A novel method for MicroRNA secondary structure prediction using a bottom-up algorithm. Proceedings of the 47th Annual Southeast Regional Conference, Clemson, South Carolina, SESSION: Information analysis, Article No. 46, ISBN: 978-1-60558-421-8 (http://doi.acm.org/10.1145/1566445.1566508) (4) Publications before Establishing My Independent Lab at the University of Kentucky and Michigan Technological University 1. Tang, G.* (2005) siRNA and miRNA: an insight into RISCs. Trends in Biochemical Sciences 30: 106-114. 2. Stepansky, A., Yao, Y., Tang, G., Galili, G. (2005) Regulation of lysine catabolism in Arabidopsis through concertedly-regulated synthesis of the two distinct gene products of the composite AtLKR/SDH locus. J. Exp. Bot. 56: 525-536 3. Mallory, A.C., Reinhart, B.J., Rhoades M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004). MicroRNA control of PHABULOSA during leaf development: importance of pairing to the miRNA 5' region. EMBO J. 23: 33563364. 4. Tang, G.* and Galili, G. (2004) Using RNAi to improve plant nutritional value: from mechanism to application. Trends in Biotechnology 22: 463-469. 5. Tang, G. and Zamore, P.D.(2004) Biochemical dissection of RNA silencing in plants, "Methods in Molecular Biology; mRNA Processing and Metabolism- Methods and Protocols" edited by Daniel R. Schoenberg , Ohio State University, Clumbus, OH, USA. HUMANA PRESS, Vol. 257: 223-243. 6. Haley, B. Tang, G. and Zamore P.D. (2003) In Vitro Analysis of RNA interference in Drosophila melanogaster. Methods 30: 330-336 7. Tang, G., B.J. Reinhart, D.P. Bartel, P.D. Zamore. (2003). A biochemical framework for RNA silencing in plants. Genes Dev 17: 49-63 8. Zhu, X, Tang, G. and Galili G. (2002) The activity of the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase enzyme of lysine catabolism is regulated by functional interaction between its two enzyme domains. J Biol Chem. 277: 49655-49661 9. Tang, G., Zhu, X., Gakiere, B., Levanony, H., Kahana, A. and Galili, G. (2002) The bifunctional LKR/SDH locus of plants also encodes a highly active monofunctional 6 Tang Updated CV lysine-ketoglutarate reductase using a polyadenylation signal located within intron. Plant Physiol.130:147 10. Galili, G., Tang, G., Zhu, X. and Gakiere, B. (2001) Lysine catabolism: a stress and development super-regulated metabolic pathway. Current Opinion in Plant Biology, 4: 261-266 11. Galili G, Tang G, Zhu X, Karchi H, Miron D, Gakiere B, and Stepansky A. (2001) Molecular genetic dissection and potential manipulation of lysine metabolism in seeds.J Plant Physiol.158: 515-520 12. Zhu, X., Tang, G., Granier, F., Boucher, D. and Galili, G. (2001) A T-DNA insertion knockout of the bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase gene elevates lysine levels in Arabidopsis seeds. Plant Physiol.126: 1539-1545 13. Tang, G., Zhu, X., Tang, X. and Galili, G. (2000) A novel composite locus encoding simultaneously two polypeptides with metabolically related but distinct functions in lysine catabolism. Plant J., 23: 195-203 14. Zhu, X., Tang, G. and Galili, G. (2000) The catabolic function of the a-amino adipic acid pathway in plants is correlated with unidirectional activity of lysine-ketoglutarate reductase, but not saccharopine dehydrogenase. Biochemical J., 351: 215-220 15. Galili, G., Tang, G., Zhu, X., Amir, R., Levanony, H., Shy, G. and Elliot M. Herman (2000) Plant seeds: an exciting model system for dissecting molecular and cellular regulation of metabolic processes. Isr. J. Plant Sci. 48: 181-187 16. Zhu, X., Tang, G. and Galili, G.(2000) Characterization of the two saccharopine dehydrogenase isozymes of lysine catabolism encoded by the single composite AtLKR/SDH locus of Arabidopsis. Plant Physiol.124: 1363-1371 17. Miron, D., Tang, G., Karchi, H., Schupper, A., Ben-Yaacob, S. and Galili,G.(1998) Regulation of Lysine Catabolism in Plants. A. Altman et al. (eds.), Plant Biotechnology and In Vitro Biology in the 21st Century, Kluwer Academic Publishers.311-314 18. Tang, G., Zhu-Shimoni,J.X., Amir,R., Zchori,I.B. and Galili,G.(1997a) Cloning and expression of an Arabidopsis thaliana cDNA encoding a monofunctional aspartate kinase homologous to the lysine-sensitive enzyme of Escherichia coli. Plant Mol. Biol. 34: 287-294 19. Tang, G., Miron, D., Zhu-Shimoni, JX., and Galili, G.(1997b) Regulation of Lysine Catabolism through Lysine-Ketoglutarate Reductase and Saccharopine Dehydrogenase in Arabidopsis. The Plant Cell, 9:1305-1316 SERVICE AND RECOGNITION Panel member US Department of Agriculture (USDA) Panel: Plant Biology: Gene Expression and Genetic Diversity 2007. International Meeting Convener, Gene Expression (Non-coding RNAs and Regulated Transgene Expression). 2007 In Vitro Biology Meeting. Indianapolis, IN. June 9-13, 2007 7 Tang Updated CV Keynote Speaker, 2008 “Small RNA technologies: siRNA, miRNA, target mimicry, and miRNA profiling.” International Association of Plant Biotechnology (IAPB) Conference - Canadian Section; MAY 5 - 8, 2008 University of Saskatchewan Saskatoon, Saskatchewan, Canada. Guest Editor (2008), “BBA / Gene Structure and Expression - Special Issue on MicroRNA” Associate Editor (2011-2014), Editorial Board for the International Journal “BMC Plant Biology” (Impact factor in 2009: 3.774) published by BioMed Central. Editorial Member (2011-2013), “Silence” published by BioMed Central. Journal reviewer service I have reviewed manuscripts more than once from the following journals: Proceedings of the National Academy of Sciences of the United States of America (PNAS), Nature Structural & Molecular Biology, EMBO Reports, The Plant Cell, Plant Journal, Plant Physiology, Cell Research, Journal of Biological Chemistry, Gene, Methods, Nucleic Acid Research, FEBS Letter, Trends in Plant Science, Metabolomic, Plant Biotechnology Journal, Plos Computational Biology, Plos ONE, Plos Genetics, BMC Bioinformatics, BMC Plant Biology, BMC Genomics, International J. of Bioch & Cell Bio, Journal of RNAi and Gene Silencing, BBA, EEB., RNA, J. of Plant Physiology, Plant Cell Reports, Hereditas, International Journal of Biological Sciences, Acta Physiologiae Plantarum, Journal of Experimental Botany. Ad hoc proposal reviewer service: USDA-NRI, NSF, BARD, USDA-SBIR, ISF (Israeli NSF), Ohio Plant Biotechnology Consortium, SOUTHWEST CONSORTIUM ON PLANT GENETICS AND WATER RESOURCES. Invited Presentations I. Invited talks or presentations in the USA and Canada: Plant and Animal Genome XXIII Conference, “Inactivation of microRNAs in Crop Plants Using Short Tandem Target Mimic (STTM) Technology”, Town & Country Convention Center, San Diego, California. January 10-14, 2015. Bayer CropScience LP, “Inactivation of microRNAs by STTM and Silencing Coding Gene by Artificial microRNAs”, Morrisville, NC. October 27, 2014. Plant and Animal Genome XXI Conference, “Regulation of Small RNAs by Small Tandem Target Mimic (STTM) in Plants and Animals”, Town & Country Convention Center, San Diego, California. January 12-16, 2013. 2011 Keystone Symposia Meeting, Mechanisms and Biology of Silencing, “Effective Small RNA Destruction by a Short Tandem Target Mimic through the Small RNA degrading Nucleases in Arabidopsis thaliana.” March 20-25, 2011, 8 Tang Updated CV Portola Hotel & Spa, Monterey, California, USA. 12th World Congress of the International Association for Plant Biotechnology (IAPB) and the 2010 In Vitro Biology Meeting of the Society for In Vitro Biology (SIVB) "Gene Regulation by Small RNAs and the Technological Application in Plants." Tuesday, June 8, 2010, America’s Center, 701 Convention Plaza, St. Louis, MO Dept. of Biology, University of South Dakota, Vermillion, SD "Gene Regulation by Small RNAs and the Technological Application in Plants." April 29, 2010 2010 Kriton-Hatzios symposium of Southern Section of the American Society of Plant Biologists, “The role of small RNAs in the regulation of gene expression” Title: "Gene Regulation by Small RNAs and the Technological Application in Plants." SS-ASPB, April 10-12 2010 University of Tennessee, Knoxville. 2010 Keystone Symposia Meeting, RNA Silencing Mechanisms in Plants, “Small RNA Destruction by a Novel Short Target Mimic in Arabidopsis thaliana.” Feb 21-26, 2010, Santa Fe, New Mexico, USA. Dept. of Agronomy, University of Wisconsin, Madison, “Small RNA technologies: siRNA, miRNA, target mimicry, and miRNA profiling” Feb. 23, 2009. Plant Metabolic Pathway Regulation and Drug Discovery Workshop, Plant and Animal Genome XVII Conference, “Small RNA Technologies For Functional Genomics And Metabolic Engineering: siRNA, MicroRNA, Target Mimicry, And MicroRNA Profiling”, Town & Country Convention Center, San Diego, California. January 10-14, 2009. Dept. of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK. “Profiling microRNAs in plants and animals”. Feb. 15th, 2008. Dept. of Biological Science and The Plant Science Initiative, University of Nebraska Lincoln. “Role of microRNA in Plant Development and Abiotic Stress”. Feb. 25th, 2008. International Association of Plant Biotechnology (IAPB) Conference - Canadian Section, “Small RNA technologies: siRNA, miRNA, target mimicry, and miRNA profiling” (an invited keynote talk). MAY 5 - 8, 2008 University of Saskatchewan, Saskatoon, Saskatchewan, Canada. “MicroRNA array technology and its applications”. Invited Speaker, MAY 8 - 9, 2008, Applied Biological Materials Inc. 9117 Shaughnessy St. Vancouver, BC, Canada V6P 6R9. 9 Tang Updated CV Meeting Conveners. Gene Expression (Non-coding RNAs and Regulated Transgene Expression). 2007 In Vitro Biology Meeting. Indianapolis, IN. June 913, 2007 24th SYMPOSIUM IN PLANT BIOLOGY, UNIVERSITY OF CALIFORNIA, RIVERSIDE. “MicroRNA profiling in plants and animals.” “Gene Silencing: The Biology of Small RNAs and the Epigenome” January18-20, 2007, Riverside Convention Center, Riverside, California Dept. of Plant Pathology, University of Kentucky, Lexington, KY. “Breakthrough Technologies in RNAi and microRNA and Wide Applications in Plant Research”, October 2009. UK-Alltech Nutrigenomics Alliance. Microarray Analyses: A Seminar Miniseries, “MicroRNA Macroarray Technology and its Application in Animals, Human Health, and Plants” Oct. 8th, 2007. Dept. of Biology, University of Kentucky, Lexington, KY. “microRNAs and RNAi in plants and Drosophila. October 2006. II. Invited talks or presentations in other departments at MTU: Dept. of Biological Sciences, Michigan Tech University, Houghton, MI. “Small Tandem Target Mimic (STTM): A New Tool for Investigating Small RNA” March 2, 2012 III. Invited talks or presentations in China: Guiliang Tang (Invited speaker), “Small RNA destruction by STTM and its application in plants.” Institute of Genetics and Developmental Biology Chinese Academy of Sciences. Beijing China. June 27, 2012 - June 29, 2012 Guiliang Tang (Invited speaker), “Small Tandem Target Mimic (STTM): A New Tool for Investigating Small RNA.” College of Agriculture, Henan Agricultural University, Zhengzhou, China. March 20, 2012 - March 29, 2012 Guiliang Tang (Invited speaker), “Gene regulation by small RNAs and the technological application in plants ” State Key Laboratory of Plant Physiology and Biochemistry, College of Life Sciences, China Agricultural University, Beijing, China. June. 21, 2011 Guiliang Tang (Invited speaker), “Gene regulation by small RNAs and the technological application in plants ”, School of Agricultural and Food Science, ZheJiang A & F University, China. Dec. 6, 2010 Guiliang Tang (Invited speaker), 1. “Small RNA technologies: siRNA, miRNA, target mimicry, and miRNA profiling”; 2. "Biochemical Dissection of RNAi and microRNA pathways and RISC assembly", College of Life Sciences, Northeast Normal University, China. Sept. 2-5, 2008 10 Tang Updated CV Guiliang Tang (Invited speaker), “Small RNA technologies: siRNA, miRNA, target mimicry, and miRNA profiling”. 2008 Department of Plant Pathology, Nanjing Agricultural University, China. Sept. 8, 2008 Guiliang Tang, “RNAi and microRNA: RISC assembly and miRNA atlas” Tsinghua University, China. Nov. 5, 2007. Guiliang Tang, “microRNA Technologies and Their Applications in Plants and Animals” Northwest A&F University (NWAFU), Xian, China Nov 6, 2007 Guiliang Tang, “RNAi and microRNA: RISC assembly and miRNA atlas” Northwest A&F University (NWAFU), Xian, China Nov. 7, 2007. Guiliang Tang, “RNAi and microRNA: RISC assembly and miRNA atlas” National Institute of Biological Science (NIBS), Beijing, China Nov. 9, 2007. Guiliang Tang, “RNAi and microRNA: RISC assembly and miRNA atlas” Genetics Institute, CAS, Beijing, China. Nov. 9, 2007. DEPARTMENTAL SERVICE Hatch Proposal Review Committee, University of Kentucky, 2007-2011 Social Committee Departmental Graduate Committee Departmental Greenhouse Committee Departmental Seminar Committee TEACHING AND ADVISING Graduate Students- Current Mengmeng Qiao, Ph.D. level candidate, 2013. Yiyou Gu, Ph.D. level candidate, 2012. Haiping Liu, Ph.D. level candidate, 2012. Li Chen, Ph.D. level candidate, 2012. Visiting Scholars/Students with external scholarship/fellowships- Current Aihua Sha Yongjun Wu Zhanhui Zhang Fanrong Meng Lina Shi Lei Tian Ting Peng Xining Jin Wenhui Wei Sachin Teotia Graduate Students- Completed Venugopal Mendu, Ph.D., 2008 Dissertation: “Roles of microRNAs in Plant Development and Abiotic Stress” International student recruitment: Xun Zhuang (graduate student), Liuyin Ma (graduate student), Ligang Ren (visiting scholar), Yanhong Bai (visiting scholar), Yanming Wei 11 Tang Updated CV (visiting scholar), Yiyou Gu (visiting scholar), Mengmeng Qiao (visiting scholar). Postdoctoral Scholars- Advised Xiaoqing Tang (2005-2008) Wangxia Wang (2006-2007) Xiaohu Tang (2007-2008) Qi-Jun Chen (2008-2009) Jun Yan (2010-present) Visiting Students- Advised Ligang Ren (2008-2010, Northwest A&F University) Xiaoran Wang (2011-2012, Northeast Normal University) Ting Peng (2013-present, Henan Agricultural University) Lei Tian (2013-present, Henan Agricultural University) Lina Shi (2013-present, Henan Agricultural University) Yiyou Gu (2008-2010, Nanjing Agricultural University) Shangjin Pan (2009-2011, Anhui University) Eric S Riedeman (For two weeks in 2009, University of Wisconsin) Wenjun Kang (2009-2011,Hefei University of Technology) Tian Yang (For three months in 2009, University of Pennsylvania) Visiting Scholars- Advised Xiaoyun Jia, (2008- present) Undergraduate Students- Current Michael L. Harrington Undergraduate Students- Advised Richard Henry Sullivan Ifedayo Olutomi Awe Cody Damm Jessica Ina Houtz High school Students- Advised Julia Li Graduate Students- Trained in RNAi Research Techniques Bobby Gaffney (USDA, UK) Kate Martin (Plant Pathology, UK) Ricky Lewis (Plant and Soil Sciences, UK) Rae Jeong (Plant Pathology, UK) Lyssa Scheuneman (Biological Science, MTU) Graduate Students- Committees Ricky Lewis (Plant and Soil Sciences, UK) Rae Jeong (Plant Pathology, UK) Yiping Mao (Biological Sciences, MTU) Ramkumar Mohan (Biological Sciences, MTU) 12 Tang Updated CV Ni Fan (Chemistry, MTU) Teaching Activities My primary teaching responsibility at MTU is for the Biochemistry I (BL4010, 3 credits) and Biochemistry Orientation (BL1800, 1 credit). Starting from September 2012, I have taught BL4010 for more than 40 undergraduate and graduate students and BL1800 for 15 undergraduates. These courses are very important to lay a foundation for the students for further study and advanced career training. I have also team taught the graduate course, Advanced Molecular Biology (BMB 6020, 3 credits) and undergraduate students Molecular Biology Seminar (BL1900, 1 credit) in Spring 2013. From Spring 2014, I will start teaching Biochemistry Techniques I (BL4820, 2 credits lab course) for undergraduate and graduate students. Course BL4010 (3 credits) BL1800 (1 credits) BMB6020 (3 credits) BL1900 (1 credits) BL4010 (3 credits) BL1800 (1 credits) BL4820 (3 credits) Fall, 2012 45 Overall Evaluation1 Course Instruction 4.3 4.24 Fall, 2012 15 4.0 3.83 Spring, 2013 13 3.9 3.58 Spring, 2013 6 4.1 4.00 Fall, 2013 33 4.1 3.96 Fall, 2013 9 4.2 4.50 Spring, 2014 13 TO BE EVALUATED Semester/Year Student number In addition to Biochemistry I (BL4010), Biochemistry Orientation (BL1800), Advanced Molecular Biology (BMB 6020), and Molecular Biology Semiar (BL1900), I have previously taught the graduate course “Experimental Course: RNAi and microRNA” (PLS 597a). The main emphasis of the course is to help senior undergraduate and beginning graduate students apply cutting-edge RNAi and microRNA technology in biochemistry, biology, and plant science to understand gene silencing for the purpose of functional genomics. Upon other faculty members’ request, I also provided guest lectures in Genomics (PLS 597b) and Introduction to Agricultural Biotechnology (ABT 101). Course Lab: RNAi & Semester/Year Spring 2007 Student number 7 Overall Evaluation1 Course Instruction 3.8 3.8 13 Tang Updated CV MicroRNA (PLS597a) Plant Genomics Spring 2007 >7 N/E2 N/E (3 lectures) (PLS597b) ABT101 Spring 2007 >30 N/E N/E (1 lecture)_________________________________________________________________ Lab: RNAi & Spring 2008 5 N/E N/E MicroRNA (PLS597a) Lab: RNAi & Spring 2010 2 N/E N/E MicroRNA (PLS597a) Note: 1 rating scale: 1 poor - 4 excellent; 2 not evaluated due to student numbers less than the minimum required for a formal TCE. PROFESSIONAL DEVELOPMENT Professional meeting attended 1. 24th SYMPOSIUM IN PLANT BIOLOGY, UNIVERSITY OF CALIFORNIA, RIVERSIDE. “Gene Silencing: The Biology of Small RNAs and the Epigenome” January18-20, 2007 2. International Association of Plant Biotechnology (IAPB) Conference - Canadian Section; MAY 5 - 8, 2008 3. Annual USDA-NRI Report Meetings (2007, 2008, 2009) at Washington D.C. 4. Plant Metabolic Pathway Regulation and Drug Discovery Workshop, Plant and Animal Genome XVII Conference, Town & Country Convention Center, San Diego, California. January 10-14, 2009. 5. KEYSTONE SYMPOSIA on Molecular and Cellular Biology, The Biology of RNA Silencing, April 25-30, 2009, Fairmont Empress Victoria, Victoria, British Columbia, Canada 6. KEYSTONE SYMPOSIA on Molecular and Cellular Biology, The Biology of RNA Silencing, April 25-30, 2009, Fairmont Empress Victoria, Victoria, British Columbia, Canada 7. KEYSTONE SYMPOSIA, RNA Silencing Mechanisms in Plants, Feb 21-16, 2010, Hilton Santa Fe/Historic Plaza, Santa Fe, New Mexico, USA 8. KEYSTONE SYMPOSIA, Mechanisms and Biology of Silencing, March 20-25, 2011, Portola Hotel & Spa, Monterey, California, USA. 9. Graduate training course in Small RNA Biology, June 29-July 30, 2012, Zhengzhou, China. 14 Tang Updated CV 10. Plant and Animal Genome XXI Conference, Town & Country Convention Center, San Diego, California. January 12-16, 2013. ACADEMIC MEMBERSHIPS RNA Society American Society of Plant Biologist REFERENCES 1. Blake Meyers, Professor/Chair Department of Plant and Soil Sciences University of Delaware Phone: 302-831-3418 E-mail: meyers@dbi.udel.edu 2. Xuemei Chen, Professor/HHMI investigator, Department of Botany and Plant Sciences University of California Riverside, CA 92521 Phone: 951 827-3988 FAX: 951 827-4437 Email: xuemei.chen@ucr.edu 3. Jian-Kang Zhu, Professor Purdue University Horticulture and Landscape Architecture 625 Agriculture Mall Drive West Lafayette, IN 47907 Phone 765-496-7601 Fax 765-494-0391 Email: jkzhu@purdue.edu 4. Harold Trick, Professor Department of Plant Pathology 3729 Throckmorton Plant Sciences Center Kansas State University, Manhattan, KS 66506-5502 (785) 532-1426 (785)532-5692 (FAX) Email: HNT@ksu.edu 5. Robert Last, Barnett Rosenberg Professor Department of Biochemistry & Molecular Biology Michigan State University 301 Biochemistry Building East Lansing, MI 48824 Office: (517) 432-3278 Lab: (517) 432-3277 15 Tang Updated CV Email: lastr@msu.edu 16